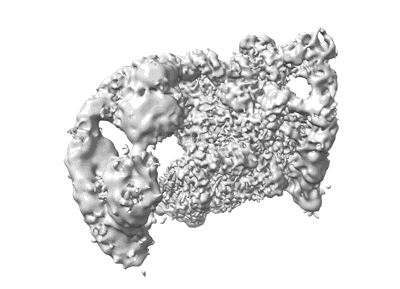
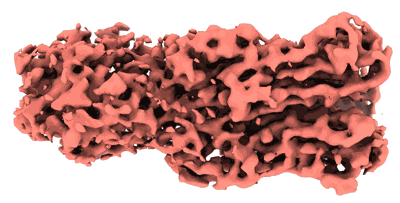
Cryo-EM map of the intact shell of alpha-carboxysome from Prochlorococcus MED4
Zhou RQ, Jiang YL, Li H, Hou P, Kong WW, Deng JX, Chen Y, Zhou CZ, Zeng Q
Nat Plants (2024) 10 pp. 661-672 [ DOI: doi:10.1038/s41477-024-01660-9 Pubmed: 38589484 ]
- The scaffolding protein csos2 (Protein from Prochlorococcus)
- The shell hexamer csos1 (Protein)
- Alpha-carboxysome shell (39 MDa, Complex from Prochlorococcus sp. MED4)
- The shell pentamer csos4a (Protein from Prochlorococcus)
Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 8wxb
Q-score: 0.337
Zhou RQ, Jiang YL, Li H, Hou P, Kong WW, Deng JX, Chen Y, Zhou CZ, Zeng Q
Nat Plants (2024) 10 pp. 661-672 [ DOI: doi:10.1038/s41477-024-01660-9 Pubmed: 38589484 ]
- Major carboxysome shell protein csos1 (10 kDa, Protein from Prochlorococcus sp. MED4)
- Carboxysome shell vertex protein csos4a (9 kDa, Protein from Prochlorococcus sp. MED4)
- The shell vertex of alpha-carboxysome (3 MDa, Complex from Prochlorococcus sp. MED4)
- Carboxysome assembly protein csos2 (82 kDa, Protein from Prochlorococcus sp. MED4)
Cryo-EM map of the intact alpha-carboxysome from Prochlorococcus MED4
Zhou RQ, Jiang YL, Li H, Hou P, Kong WW, Deng JX, Chen Y, Zhou CZ, Zeng Q
Nat Plants (2024) 10 pp. 661-672 [ DOI: doi:10.1038/s41477-024-01660-9 Pubmed: 38589484 ]
- Alpha-carboxysome (97 MDa, Complex from Prochlorococcus)
- The scaffolding protein csos2 (Protein from Prochlorococcus)
- The shell pentamer csos4b (Protein from Prochlorococcus)
- Rubisco (Protein from Prochlorococcus)
- The shell pentamer csos4a (Protein from Prochlorococcus)
- The shell trimer csos1d (Protein from Prochlorococcus)
- The carbonic anhydrase csosca (Protein from Prochlorococcus)
- The shell hexamer csos1 (Protein from Prochlorococcus)
Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 8h3v
Q-score: 0.191
Han SJ, Jiang YL, You LL, Shen LQ, Wu X, Yang F, Cui N, Kong WW, Sun H, Zhou K, Meng HC, Chen ZP, Chen Y, Zhang Y, Zhou CZ
Nat Struct Mol Biol (2024) 31 pp. 293-299 [ DOI: doi:10.1038/s41594-023-01149-7 Pubmed: 38177666 ]
- Dna (125-mer) (38 kDa, DNA from Anabaena)
- Dna-directed rna polymerase subunit alpha (26 kDa, Protein from Anabaena)
- Dna-directed rna polymerase subunit gamma (70 kDa, Protein from Anabaena)
- Rna polymerase sigma factor siga (45 kDa, Protein from Anabaena)
- Dna (125-mer) (38 kDa, DNA from Anabaena)
- Full transcription activation complex with two activators ntca and ntcb (718 kDa, Complex from Anabaena)
- Dna-directed rna polymerase subunit beta (126 kDa, Protein from Anabaena)
- Dna-directed rna polymerase subunit omega (9 kDa, Protein from Anabaena)
- Ntca (24 kDa, Protein from Anabaena)
- Ntcb (34 kDa, Protein from Anabaena)
- Dna-directed rna polymerase subunit beta' (147 kDa, Protein from Anabaena)
Cryo-EM structure of the transcription activation complex NtcA-TAC
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8h40
Q-score: 0.353
Han SJ, Jiang YL, You LL, Shen LQ, Wu X, Yang F, Cui N, Kong WW, Sun H, Zhou K, Meng HC, Chen ZP, Chen Y, Zhang Y, Zhou CZ
Nat Struct Mol Biol (2024) 31 pp. 293-299 [ DOI: doi:10.1038/s41594-023-01149-7 Pubmed: 38177666 ]
- Dna (125-mer) (38 kDa, DNA from Anabaena)
- Dna-directed rna polymerase subunit alpha (26 kDa, Protein from Anabaena)
- Dna-directed rna polymerase subunit gamma (70 kDa, Protein from Anabaena)
- Rna polymerase sigma factor siga (45 kDa, Protein from Anabaena)
- Transcription activation complex with the global regulator ntca (580 kDa, Complex from Anabaena)
- Dna (125-mer) (38 kDa, DNA from Anabaena)
- Dna-directed rna polymerase subunit beta (126 kDa, Protein from Anabaena)
- Dna-directed rna polymerase subunit omega (9 kDa, Protein from Anabaena)
- Ntca (24 kDa, Protein from Anabaena)
- Dna-directed rna polymerase subunit beta' (147 kDa, Protein from Anabaena)
Resolution: 3.66 Å
EM Method: Single-particle
Fitted PDBs: 8hdr
Q-score: 0.445
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ DOI: doi:10.1073/pnas.2213727120 Pubmed: 36656854 ]
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Pam3 tube protein (15 kDa, Protein from uncultured cyanophage)
- Pam3 sheath protein (41 kDa, Protein from uncultured cyanophage)
- Pam3 terminator protein (17 kDa, Protein from uncultured cyanophage)
- Pam3 connector protein (12 kDa, Protein from uncultured cyanophage)
- Pam3 adaptor protein (13 kDa, Protein from uncultured cyanophage)
Resolution: 3.96 Å
EM Method: Single-particle
Fitted PDBs: 7yfw
Q-score: 0.413
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ DOI: doi:10.1073/pnas.2213727120 Pubmed: 36656854 ]
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Pam3 fiber proreins (27 kDa, Protein from uncultured cyanophage)
Cyanophage Pam3 baseplate proteins
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 7yfz
Q-score: 0.528
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ DOI: doi:10.1073/pnas.2213727120 Pubmed: 36656854 ]
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Pam3 baseplate wedge gp23 (26 kDa, Protein from uncultured cyanophage)
- Pam3 tube initiator gp17 (19 kDa, Protein from uncultured cyanophage)
- Pam3 hub gp19 (25 kDa, Protein from uncultured cyanophage)
- Pam3 plug gp18 (11 kDa, Protein from uncultured cyanophage)
- Pam3 sheath initiator gp21 (13 kDa, Protein from uncultured cyanophage)
- Pam3 baseplate wedge gp22 (32 kDa, Protein from uncultured cyanophage)
- Pam3 spike gp20 (23 kDa, Protein from uncultured cyanophage)