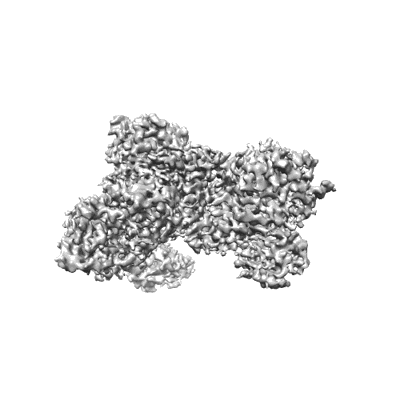
Cryo-EM structure of human DNMT3B homo-tetramer (form I)
Resolution: 3.04 Å
EM Method: Single-particle
Fitted PDBs: 8eih
Q-score: 0.493
Lu J, Fang J, Zhu H, Liang KL, Khudaverdyan N, Song J
Nucleic Acids Res (2023) 51 pp. 12476-12491 [ DOI: doi:10.1093/nar/gkad972 Pubmed: 37941146 ]
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (cytosine-5)-methyltransferase 3b (Complex from Homo sapiens)
- Dna (cytosine-5)-methyltransferase 3b (73 kDa, Protein from Homo sapiens)
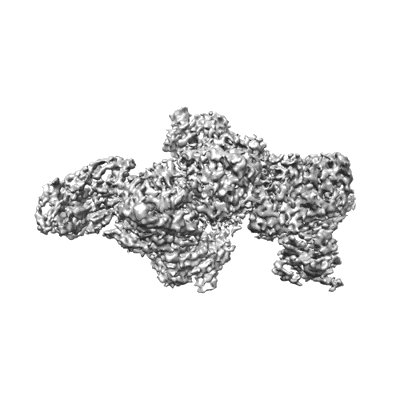
Cryo-EM structure of human DNMT3B homo-tetramer (form II)
Resolution: 3.12 Å
EM Method: Single-particle
Fitted PDBs: 8eii
Q-score: 0.44
Lu J, Fang J, Zhu H, Liang KL, Khudaverdyan N, Song J
Nucleic Acids Res (2023) 51 pp. 12476-12491 [ DOI: doi:10.1093/nar/gkad972 Pubmed: 37941146 ]
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (cytosine-5)-methyltransferase 3b (Complex from Homo sapiens)
- Dna (cytosine-5)-methyltransferase 3b (73 kDa, Protein from Homo sapiens)
Cryo-EM structure of human DNMT3B homo-trimer
Resolution: 3.34 Å
EM Method: Single-particle
Fitted PDBs: 8eij
Q-score: 0.426
Lu J, Fang J, Zhu H, Liang KL, Khudaverdyan N, Song J
Nucleic Acids Res (2023) 51 pp. 12476-12491 [ DOI: doi:10.1093/nar/gkad972 Pubmed: 37941146 ]
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (cytosine-5)-methyltransferase 3b (Complex from Homo sapiens)
- Dna (cytosine-5)-methyltransferase 3b (73 kDa, Protein from Homo sapiens)
Cryo-EM structure of human DNMT3B homo-hexamer
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 8eik
Q-score: 0.383
Lu J, Fang J, Zhu H, Liang KL, Khudaverdyan N, Song J
Nucleic Acids Res (2023) 51 pp. 12476-12491 [ DOI: doi:10.1093/nar/gkad972 Pubmed: 37941146 ]
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (cytosine-5)-methyltransferase 3b (Complex from Homo sapiens)
- Dna (cytosine-5)-methyltransferase 3b (73 kDa, Protein from Homo sapiens)
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs
Resolution: 3.49 Å
EM Method: Single-particle
Fitted PDBs: 7chh
Q-score: -0.009
Du S, Cao Y, Zhu Q, Yu P, Qi F, Wang G, Du X, Bao L, Deng W, Zhu H, Liu J, Nie J, Zheng Y, Liang H, Liu R, Gong S, Xu H, Yisimayi A, Lv Q, Wang B, He R, Han Y, Zhao W, Bai Y, Qu Y, Gao X, Ji C, Wang Q, Gao N, Huang W, Wang Y, Xie XS, Su XD, Xiao J, Qin C
Cell (2020) 183 pp. 1013-1023.e13 [ Pubmed: 32970990 DOI: doi:10.1016/j.cell.2020.09.035 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Complex of the sars-cov-2 s-6p with bd-368-2 fabs (Complex)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Bd-368-2 fab light chain (23 kDa, Protein from Homo sapiens)
- Bd-368-2 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Bd-368-2 fabs (Complex from Homo sapiens)
Cryo-EM Structure of Isomeric Molluscan Hemocyanin Type 1 triggered by viral infection