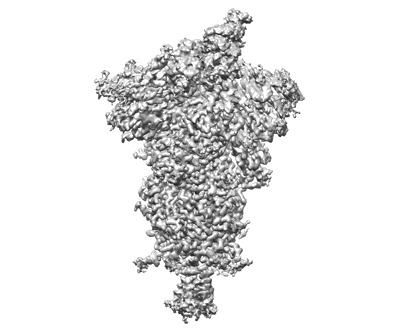
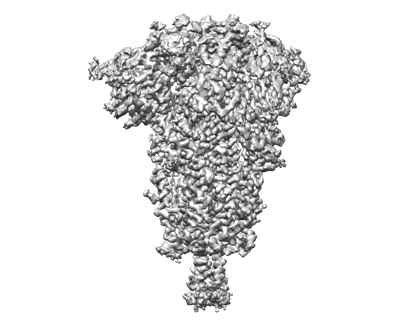
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8fdw
Q-score: 0.327
Shi W, Cai Y, Zhu H, Peng H, Voyer J, Rits-Volloch S, Cao H, Mayer ML, Song K, Xu C, Lu J, Zhang J, Chen B
Nature (2023) 619 pp. 403-409 [ Pubmed: 37285872 DOI: doi:10.1038/s41586-023-06273-4 ]
- Spike protein s2 (68 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sras-cov-2 postfusion spike protein in nanodisc (210 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Alpha-d-mannopyranose (180 Da, Ligand)
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7n1q
Q-score: 0.472
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 3.11 Å
EM Method: Single-particle
Fitted PDBs: 7n1t
Q-score: 0.458
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 3.14 Å
EM Method: Single-particle
Fitted PDBs: 7n1u
Q-score: 0.44
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 3.21 Å
EM Method: Single-particle
Fitted PDBs: 7n1v
Q-score: 0.445
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 3.33 Å
EM Method: Single-particle
Fitted PDBs: 7n1w
Q-score: 0.434
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 7n1x
Q-score: 0.364
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 7n1y
Q-score: 0.321
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]