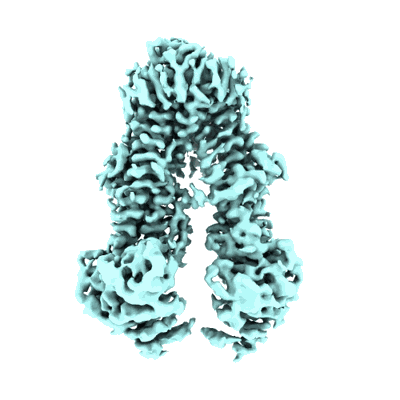
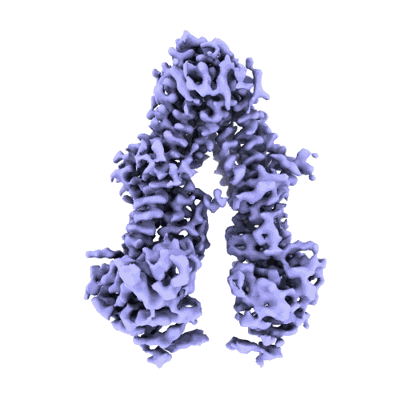
Cryo-EM structure of Cas5-HNH Cascade bound with dsDNA
Resolution: 2.93 Å
EM Method: Single-particle
Fitted PDBs: 9jxs
Q-score: 0.446
Liu YN, Wang L, Zhang H, Zhu H
To Be Published
- Crispr system cascade subunit casc (41 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Cas5-hnh cascade complex bound with dna (400 MDa, Complex from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Non-target dna (16 kDa, DNA from Escherichia coli)
- Dna (54-mer) (16 kDa, DNA from Escherichia coli)
- Rna (61-mer) (19 kDa, RNA from Escherichia coli)
- Crispr system cascade subunit casd (43 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Magnesium ion (24 Da, Ligand)
- Crispr-associated protein cse2 (crispr_cse2) (20 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Crispr-associated protein cse1 (crispr_cse1) (60 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Crispr-associated endoribonuclease cse3 (31 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
Cryo-EM structure of Cas5-HNH Cascade bound with sDNA, Conf1
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8zp7
Q-score: 0.517
Liu YN, Wang L, Zhang H, Zhu H
To Be Published
- Crispr system cascade subunit casc (41 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Dna (60-mer) (18 kDa, DNA from Escherichia coli)
- Rna (61-mer) (19 kDa, RNA from Escherichia coli)
- Crispr-associated protein cse1 (crispr_cse1) (62 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Crispr system cascade subunit casd (43 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Magnesium ion (24 Da, Ligand)
- Cas5-hnh cascade (400 MDa, Complex from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Crispr-associated protein cse2 (crispr_cse2) (20 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
- Crispr-associated endoribonuclease cse3 (31 kDa, Protein from Candidatus Cloacimonetes bacterium ADurb.Bin088)
Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 monomer state
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 8vh4
Q-score: 0.333
Li X, Zhu H, Huang BT, Li X, Kim H, Tan H, Zhang Y, Choi I, Peng J, Xu P, Sun J, Yue Z
Nat Commun (2024) 15 pp. 8434-8434 [ Pubmed: 39343966 DOI: doi:10.1038/s41467-024-52723-6 ]
- Leucine-rich repeat serine/threonine-protein kinase 2 (286 kDa, Protein from Homo sapiens)
- Rab12-lrrk2 (Complex from Homo sapiens)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Ras-related protein rab-12 (20 kDa, Protein from Homo sapiens)
- Phosphoaminophosphonic acid-guanylate ester (522 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 dimer state
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8vh5
Q-score: 0.248
Li X, Zhu H, Huang BT, Li X, Kim H, Tan H, Zhang Y, Choi I, Peng J, Xu P, Sun J, Yue Z
Nat Commun (2024) 15 pp. 8434-8434 [ Pubmed: 39343966 DOI: doi:10.1038/s41467-024-52723-6 ]
- Leucine-rich repeat serine/threonine-protein kinase 2 (286 kDa, Protein from Homo sapiens)
- Rab12-lrrk2 (Complex from Homo sapiens)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Ras-related protein rab-12 (20 kDa, Protein from Homo sapiens)
- Phosphoaminophosphonic acid-guanylate ester (522 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
Structure of HKU1B RBD with TMPRSS2
Gao X, Zhu K, Wang L, Shang K, Hua L, Qin B, Zhu H, Ding W, Cui S
Cell Discov (2024) 10 pp. 84-84 [ Pubmed: 39112468 DOI: doi:10.1038/s41421-024-00717-5 ]
- Spike protein s1 (31 kDa, Protein from Candidatus Accumulibacter adiacens)
- Hku1b-rbd with tmprss2 receptor (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Transmembrane protease serine 2 (42 kDa, Protein from Homo sapiens)
Cryo-EM structure of succinate receptor SUCR1 bound to compound 31
Resolution: 2.48 Å
EM Method: Single-particle
Fitted PDBs: 8ykv
Q-score: 0.585
Li C, Liu H, Li J, He X, Zhu H, Fu W, Xu HE
Cell Res (2024) 34 pp. 594-596 [ Pubmed: 38834763 DOI: doi:10.1038/s41422-024-00984-7 ]
- Cryo-em structure of succinate receptor sucr1 bound to succinic acid (160 kDa, Complex from Homo sapiens)
- Antibody fragment scfv16 (26 kDa, Protein from synthetic construct)
- Succinate receptor 1 (38 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Bos taurus)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- (2~{s})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid (398 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Rattus norvegicus)
Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid
Resolution: 2.75 Å
EM Method: Single-particle
Fitted PDBs: 8ykw
Q-score: 0.553
Li C, Liu H, Li J, He X, Zhu H, Fu W, Xu HE
Cell Res (2024) 34 pp. 594-596 [ Pubmed: 38834763 DOI: doi:10.1038/s41422-024-00984-7 ]
- Succinic acid (118 Da, Ligand)
- Cryo-em structure of succinate receptor sucr1 bound to succinic acid (160 kDa, Complex from Homo sapiens)
- Antibody fragment scfv16 (26 kDa, Protein from synthetic construct)
- Succinate receptor 1 (38 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Bos taurus)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Rattus norvegicus)
Cryo-EM structure of succinate receptor SUCR1 bound to maleic acid
Resolution: 2.69 Å
EM Method: Single-particle
Fitted PDBs: 8ykx
Q-score: 0.563
Li C, Liu H, Li J, He X, Zhu H, Fu W, Xu HE
Cell Res (2024) 34 pp. 594-596 [ Pubmed: 38834763 DOI: doi:10.1038/s41422-024-00984-7 ]
- Cryo-em structure of succinate receptor sucr1 bound to succinic acid (160 kDa, Complex from Homo sapiens)
- Antibody fragment scfv16 (26 kDa, Protein from synthetic construct)
- Succinate receptor 1 (38 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Bos taurus)
- Maleic acid (116 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Rattus norvegicus)
Structure of the wild-type Arabidopsis ABCB19 in the brassinolide-bound state
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8wom
Q-score: 0.451
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
Structure of the wild-type Arabidopsis ABCB19 in the brassinolide and AMP-PNP bound state
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 8woo
Q-score: 0.401
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
- Abc transporter b family member 19 (136 kDa, Protein from Arabidopsis thaliana)
- Purified wild-type abcb19 protein in complex with brassinolide and amp-pnp (Cellular component from Arabidopsis thaliana)
- Magnesium ion (24 Da, Ligand)
- Brassinolide (480 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)