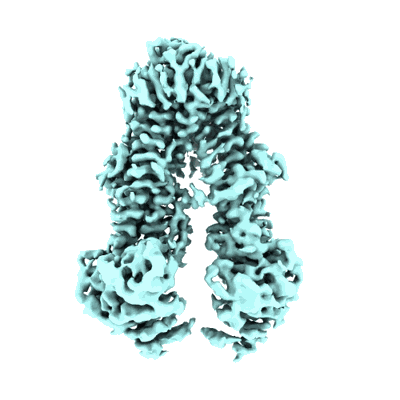
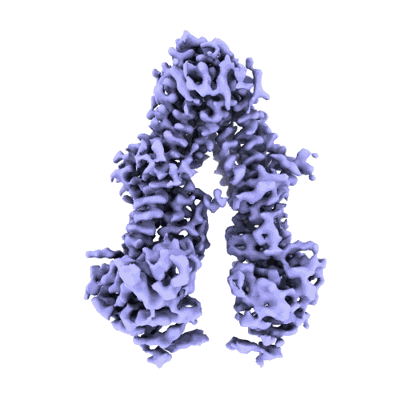
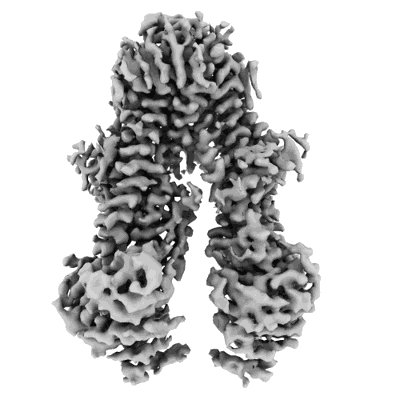
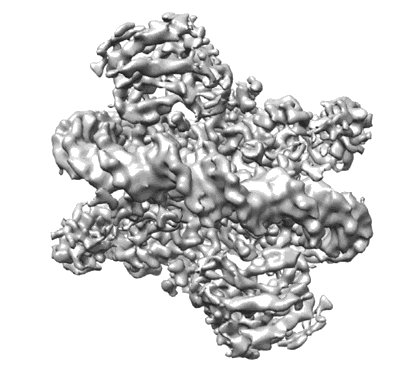
Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 monomer state
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 8vh4
Q-score: 0.333
Li X, Zhu H, Huang BT, Li X, Kim H, Tan H, Zhang Y, Choi I, Peng J, Xu P, Sun J, Yue Z
Nat Commun (2024) 15 pp. 8434-8434 [ Pubmed: 39343966 DOI: doi:10.1038/s41467-024-52723-6 ]
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Rab12-lrrk2 (Complex from Homo sapiens)
- Leucine-rich repeat serine/threonine-protein kinase 2 (286 kDa, Protein from Homo sapiens)
- Phosphoaminophosphonic acid-guanylate ester (522 Da, Ligand)
- Ras-related protein rab-12 (20 kDa, Protein from Homo sapiens)
Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 dimer state
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8vh5
Q-score: 0.248
Li X, Zhu H, Huang BT, Li X, Kim H, Tan H, Zhang Y, Choi I, Peng J, Xu P, Sun J, Yue Z
Nat Commun (2024) 15 pp. 8434-8434 [ Pubmed: 39343966 DOI: doi:10.1038/s41467-024-52723-6 ]
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Rab12-lrrk2 (Complex from Homo sapiens)
- Leucine-rich repeat serine/threonine-protein kinase 2 (286 kDa, Protein from Homo sapiens)
- Phosphoaminophosphonic acid-guanylate ester (522 Da, Ligand)
- Ras-related protein rab-12 (20 kDa, Protein from Homo sapiens)
Structure of the wild-type Arabidopsis ABCB19 in the brassinolide-bound state
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8wom
Q-score: 0.451
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
Structure of the wild-type Arabidopsis ABCB19 in the brassinolide and AMP-PNP bound state
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 8woo
Q-score: 0.401
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
- Abc transporter b family member 19 (136 kDa, Protein from Arabidopsis thaliana)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Purified wild-type abcb19 protein in complex with brassinolide and amp-pnp (Cellular component from Arabidopsis thaliana)
- Brassinolide (480 Da, Ligand)
Structure of the Arabidopsis E529Q/E1174Q ABCB19 in the ATP bound state
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8wp0
Q-score: 0.464
Ying W, Wang Y, Wei H, Luo Y, Ma Q, Zhu H, Janssens H, Vukasinovic N, Kvasnica M, Winne JM, Gao Y, Tan S, Friml J, Liu X, Russinova E, Sun L
Science (2024) 383 pp. eadj4591-eadj4591 [ DOI: doi:10.1126/science.adj4591 Pubmed: 38513023 ]
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Abc transporter b family member 19 (136 kDa, Protein from Arabidopsis thaliana)
- Purified e529q/e1174q mutant abcb19 protein in complex with atp (Cellular component from Arabidopsis thaliana)
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 7yrg
Q-score: 0.227
Huang L, Wang Y, Long H, Zhu H, Wen Z, Zhang L, Zhang W, Guo Z, Wang L, Tang F, Hu J, Bao K, Zhu P, Li G, Zhou Z
Mol Cell (2023) 83 pp. 2884-2895.e7 [ Pubmed: 37536340 DOI: doi:10.1016/j.molcel.2023.07.001 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Histone (Complex from Homo sapiens)
- Histone h2b 1.1 (10 kDa, Protein from Xenopus laevis)
- Histone methyltransferase and nucleosome complex (Complex)
- S-adenosylmethionine (398 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (146-mer) (45 kDa, DNA from Homo sapiens)
- Histone h3.2 (12 kDa, Protein from Xenopus laevis)
- Histone,suv420h1 (Complex from Xenopus laevis)
- Histone h2a.z (12 kDa, Protein from Homo sapiens)
- [histone h4]-n-methyl-l-lysine20 n-methyltransferase kmt5b (32 kDa, Protein from Homo sapiens)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7yrd
Q-score: 0.484
Huang L, Wang Y, Long H, Zhu H, Wen Z, Zhang L, Zhang W, Guo Z, Wang L, Tang F, Hu J, Bao K, Zhu P, Li G, Zhou Z
Mol Cell (2023) 83 pp. 2884-2895.e7 [ Pubmed: 37536340 DOI: doi:10.1016/j.molcel.2023.07.001 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (10 kDa, Protein from Xenopus laevis)
- S-adenosylmethionine (398 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (146-mer) (45 kDa, DNA from Homo sapiens)
- Histone methyltransferase and nucleosome complex (Complex from Homo sapiens)
- Histone h3.2 (12 kDa, Protein from Xenopus laevis)
- Histone h2a.z (12 kDa, Protein from Homo sapiens)
- [histone h4]-n-methyl-l-lysine20 n-methyltransferase kmt5b (32 kDa, Protein from Homo sapiens)
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8fdw
Q-score: 0.327
Shi W, Cai Y, Zhu H, Peng H, Voyer J, Rits-Volloch S, Cao H, Mayer ML, Song K, Xu C, Lu J, Zhang J, Chen B
Nature (2023) 619 pp. 403-409 [ Pubmed: 37285872 DOI: doi:10.1038/s41586-023-06273-4 ]
- Spike protein s2 (68 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sras-cov-2 postfusion spike protein in nanodisc (210 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Alpha-d-mannopyranose (180 Da, Ligand)
Resolution: 3.88 Å
EM Method: Single-particle
Fitted PDBs: 7wp8
Q-score: 0.441
Wu Y, Wang S, Zhang Y, Yuan L, Zheng Q, Wei M, Shi Y, Wang Z, Ma J, Wang K, Nie M, Xiao J, Huang Z, Chen P, Guo H, Lan M, Xu J, Hou W, Hong Y, Chen D, Sun H, Xiong H, Zhou M, Liu C, Guo W, Guo H, Gao J, Gan C, Li Z, Zhang H, Wang X, Li S, Cheng T, Zhao Q, Chen Y, Wu T, Zhang T, Zhang J, Cao H, Zhu H, Yuan Q, Guan Y, Xia N
Cell Host Microbe (2022) 30 pp. 1732-1744.e7 [ Pubmed: 36323313 DOI: doi:10.1016/j.chom.2022.10.011 ]
- 83h7 heavy chain (13 kDa, Protein from Mus musculus)
- Cryo-em structure of sars-cov-2 recombinant spike protein stfk1628x in complex with three neutralizing antibodies (Complex from Mus musculus)
- Neutralizing antibodies (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2b4 light chain (12 kDa, Protein from Mus musculus)
- Spike protein stfk1628x (Complex)
- 85f7 heavy chain (13 kDa, Protein from Mus musculus)
- 85f7 light chain (11 kDa, Protein from Mus musculus)
- 83h7 light chain (11 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (20 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2b4 heavy chain (13 kDa, Protein from Mus musculus)
Resolution: 3.81 Å
EM Method: Single-particle
Fitted PDBs: 7wp6
Q-score: 0.438
Wu Y, Wang S, Zhang Y, Yuan L, Zheng Q, Wei M, Shi Y, Wang Z, Ma J, Wang K, Nie M, Xiao J, Huang Z, Chen P, Guo H, Lan M, Xu J, Hou W, Hong Y, Chen D, Sun H, Xiong H, Zhou M, Liu C, Guo W, Guo H, Gao J, Gan C, Li Z, Zhang H, Wang X, Li S, Cheng T, Zhao Q, Chen Y, Wu T, Zhang T, Zhang J, Cao H, Zhu H, Yuan Q, Guan Y, Xia N
Cell Host Microbe (2022) 30 pp. 1732-1744.e7 [ Pubmed: 36323313 DOI: doi:10.1016/j.chom.2022.10.011 ]
- 83h7 heavy chain (13 kDa, Protein from Mus musculus)
- 36h6 light chain (12 kDa, Protein from Mus musculus)
- Spike protein stfk (Complex)
- Neutralizing antibodies (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s1 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 85f7 heavy chain (13 kDa, Protein from Mus musculus)
- 85f7 light chain (11 kDa, Protein from Mus musculus)
- 36h6 heavy chain (13 kDa, Protein from Mus musculus)
- 83h7 light chain (11 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cryo-em structure of sars-cov-2 recombinant spike protein stfk in complex with three neutralizing antibodies (Complex from Mus musculus)