The tetramer Structure of DSR2 alone
Zhu K, Shang K, Wang L, Yu X, Hua L, Zhang W, Qin B, Wang J, Gao X, Zhu H, Cui S
Commun Biol (2025) 8 pp. 297-297 [ Pubmed: 39994439 DOI: doi:10.1038/s42003-025-07743-3 ]
- The dimer of dsr2 (Complex from Bacillus subtilis A29)
- Sir2-like domain-containing protein (118 kDa, Protein from Bacillus subtilis A29)
The Dimer Structure of DSR2-SPR
Zhu K, Shang K, Wang L, Yu X, Hua L, Zhang W, Qin B, Wang J, Gao X, Zhu H, Cui S
Commun Biol (2025) 8 pp. 297-297 [ Pubmed: 39994439 DOI: doi:10.1038/s42003-025-07743-3 ]
- The dimer of dsr2 (Complex from Bacillus subtilis A29)
- Sir2-like domain-containing protein (118 kDa, Protein from Bacillus subtilis A29)
- Spr (29 kDa, Protein from Bacillus subtilis A29)
The tetramer Structure of SPR-DSR2 complex
Zhu K, Shang K, Wang L, Yu X, Hua L, Zhang W, Qin B, Wang J, Gao X, Zhu H, Cui S
Commun Biol (2025) 8 pp. 297-297 [ Pubmed: 39994439 DOI: doi:10.1038/s42003-025-07743-3 ]
- Sir2-like domain-containing protein (118 kDa, Protein from Bacillus subtilis A29)
- Spr (29 kDa, Protein from Bacillus subtilis A29)
- The tetramer of spr-dsr2 (Complex from Bacillus subtilis A29)
The Dimer Structure of DSR2-SPR with NAD
Zhu K, Shang K, Wang L, Yu X, Hua L, Zhang W, Qin B, Wang J, Gao X, Zhu H, Cui S
Commun Biol (2025) 8 pp. 297-297 [ Pubmed: 39994439 DOI: doi:10.1038/s42003-025-07743-3 ]
- Sir2-like domain-containing protein (118 kDa, Protein from Bacillus subtilis A29)
- The dimer of dsr2-spr with nad (Complex from Bacillus subtilis A29)
- Spr (29 kDa, Protein from Bacillus subtilis A29)
The tetramer Structure of DSR2-SPR with NAD
Zhu K, Shang K, Wang L, Yu X, Hua L, Zhang W, Qin B, Wang J, Gao X, Zhu H, Cui S
Commun Biol (2025) 8 pp. 297-297 [ Pubmed: 39994439 DOI: doi:10.1038/s42003-025-07743-3 ]
- Sir2-like domain-containing protein (118 kDa, Protein from Bacillus subtilis A29)
- The tetramer of dsr2-spr with nad (Complex from Bacillus subtilis A29)
- Spr (29 kDa, Protein from Bacillus subtilis A29)
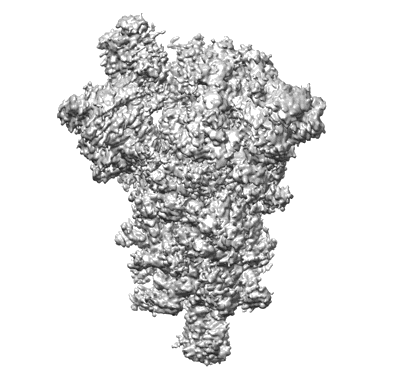
One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 7sbt
Q-score: 0.269
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ DOI: doi:10.1126/science.abl9463 Pubmed: 34698504 ]
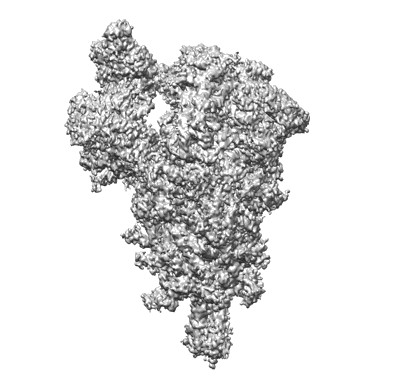
One RBD-up 2 of pre-fusion SARS-CoV-2 Delta variant spike protein
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 7sbo
Q-score: 0.266
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ DOI: doi:10.1126/science.abl9463 Pubmed: 34698504 ]
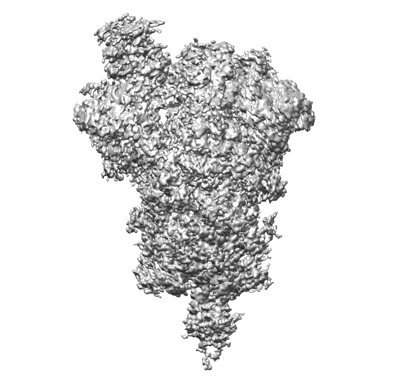
One RBD-up 2 of pre-fusion SARS-CoV-2 Kappa variant spike protein
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 7sbr
Q-score: 0.274
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ DOI: doi:10.1126/science.abl9463 Pubmed: 34698504 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 7n1x
Q-score: 0.364
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 7n1y
Q-score: 0.321
Cai Y, Zhang J, Xiao T, Lavine CL, Rawson S, Peng H, Zhu H, Anand K, Tong P, Gautam A, Lu S, Sterling SM, Walsh Jr RM, Rits-Volloch S, Lu J, Wesemann DR, Yang W, Seaman MS, Chen B
Science (2021) 373 pp. 642-648 [ DOI: doi:10.1126/science.abi9745 Pubmed: 34168070 ]