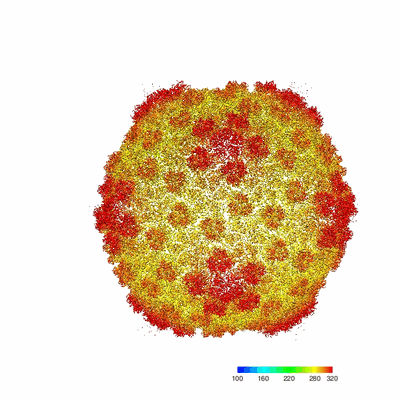
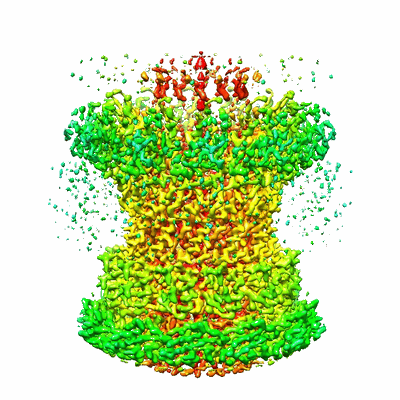Near-atomic structure of five-fold averaged PBCV-1 capsid
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 8h2i
Q-score: 0.362
Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q
Nat Commun (2022) 13 pp. 6476-6476 [ Pubmed: 36309542 DOI: doi:10.1038/s41467-022-34218-4 ]
- P1v1 (57 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- Paramecium bursaria chlorella virus 1 (Virus from Paramecium bursaria Chlorella virus 1)
- P13 (11 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P21 (141 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- Major capsid protein (mcp) (48 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P2 (63 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P15 (18 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P9 (23 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P23 (140 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- Mcpv4 (45 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P17 (9 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P5 (16 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- Mcpv1 (58 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P6 (24 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P19 (43 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P11 (23 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- Mcpv5 (45 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P3 (19 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P22 (137 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P18 (16 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P4 (20 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P12 (17 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P20 (137 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- Mcpv3 (45 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P8 (19 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- P16 (9 kDa, Protein from Paramecium bursaria Chlorella virus 1)
- Mcpv2 (55 kDa, Protein from Paramecium bursaria Chlorella virus 1)
The unexpanded head structure of phage T4
Resolution: 5.1 Å
EM Method: Single-particle
Fitted PDBs: 7vrt
Q-score: 0.202
Fang Q, Tang WC, Fokine A, Mahalingam M, Shao Q, Rossmann MG, Rao VB
PNAS (2022) 119 pp. e2203272119-e2203272119 [ Pubmed: 36161892 DOI: doi:10.1073/pnas.2203272119 ]
- Major capsid protein (56 kDa, Protein from Enterobacteria phage T4)
- Capsid vertex protein (47 kDa, Protein from Enterobacteria phage T4)
- Escherichia virus t4 (Virus from Escherichia virus T4)
The expanded head structure of phage T4
Fang Q, Tang WC, Fokine A, Mahalingam M, Shao Q, Rossmann MG, Rao VB
PNAS (2022) 119 pp. e2203272119-e2203272119 [ Pubmed: 36161892 DOI: doi:10.1073/pnas.2203272119 ]
- Small outer capsid protein (9 kDa, Protein from Enterobacteria phage T4)
- Major capsid protein (56 kDa, Protein from Enterobacteria phage T4)
- Capsid vertex protein (47 kDa, Protein from Enterobacteria phage T4)
- Escherichia virus t4 (Virus from Escherichia virus T4)
Late pre-fusion state of EEEV (pH 5.5) with localized reconstruction
Resolution: 14.1 Å
EM Method: Single-particle
Fitted PDBs: 7n69
Q-score: 0.081
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
Early pre-fusion state of EEEV (pH 5.5) with localized reconstruction
Resolution: 14.3 Å
EM Method: Single-particle
Fitted PDBs: 7n6a
Q-score: 0.069
Chen CL, Klose T, Sun C, Kim AS, Buda G, Rossmann MG, Diamond MS, Klimstra WB, Kuhn RJ
PNAS (2022) 119 pp. e2114119119-e2114119119 [ DOI: doi:10.1073/pnas.2114119119 Pubmed: 35867819 ]
The capsid of Myoviridae Phage XM1
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7kmx
Q-score: 0.541
Wang Z, Fokine A, Guo X, Jiang W, Rossmann MG, Kuhn RJ, Luo ZH, Klose T
bioRxiv (2021) [ DOI: doi:10.1101/2021.12.18.469577 ]
- Major capsid protein (35 kDa, Protein from Vibrio phage XM1)
- Vibrio phage xm1 (Virus from Vibrio phage XM1)
- Minor capsid protein (16 kDa, Protein from Vibrio phage XM1)
The Neck region of Phage XM1 (6-fold symmetry)
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7kjk
Q-score: 0.46
Wang Z, Fokine A, Guo X, Jiang W, Rossmann MG, Kuhn RJ, Luo ZH, Klose T
bioRxiv (2021) [ DOI: doi:10.1101/2021.12.18.469577 ]
- Collar spike protein (90 kDa, Protein from Vibrio phage XM1)
- Tail terminator protein (18 kDa, Protein from Vibrio phage XM1)
- Tail tube protein (15 kDa, Protein from Vibrio phage XM1)
- Vibrio phage xm1 (Virus from Vibrio phage XM1)
- Tail sheath protein (53 kDa, Protein from Vibrio phage XM1)
- Head completion protein (13 kDa, Protein from Vibrio phage XM1)
Myoviridae Phage XM1 Neck Region (12-fold)
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7kln
Q-score: 0.455
Wang Z, Fokine A, Guo X, Jiang W, Rossmann MG, Kuhn RJ, Luo ZH, Klose T
bioRxiv (2021) [ DOI: doi:10.1101/2021.12.18.469577 ]
- Head completion protein, gp1 (12 kDa, Protein from Vibrio phage XM1)
- Portal protein (47 kDa, Protein from Vibrio phage XM1)
- Vibrio phage xm1 (Virus from Vibrio phage XM1)
Baseplate Complex for Myoviridae Phage XM1
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7kh1
Q-score: 0.493
Wang Z, Fokine A, Guo X, Jiang W, Rossmann MG, Kuhn RJ, Luo ZH, Klose T
bioRxiv (2021) [ DOI: doi:10.1101/2021.12.18.469577 ]
- Baseplate organization protein, gp11 (27 kDa, Protein from Vibrio phage XM1)
- Tail sheath initiator protein, gp15 (13 kDa, Protein from Vibrio phage XM1)
- Tail sheath protein, gp6 (53 kDa, Protein from Vibrio phage XM1)
- Vibrio phage xm1 (Virus from Vibrio phage XM1)
- Baseplate wedge protein, gp17 (27 kDa, Protein from Vibrio phage XM1)
- Tail tube protein, gp7 (15 kDa, Protein from Vibrio phage XM1)
- Baseplate stabilizing protein, gp12 (13 kDa, Protein from Vibrio phage XM1)
- Baseplate wedge protein, gp16 (44 kDa, Protein from Vibrio phage XM1)
The mature Usutu SAAR-1776, Map A
Resolution: 2.42 Å
EM Method: Single-particle
Fitted PDBs: 7lcg
Q-score: 0.558
Khare B, Klose T, Fang Q, Rossmann MG, Kuhn RJ
PNAS (2021) 118 [ Pubmed: 34417300 DOI: doi:10.1073/pnas.2107408118 ]
- (7s)-4-hydroxy-n,n,n-trimethyl-9-oxo-7-[(palmitoyloxy)methyl]-3,5,8-trioxa-4-phosphahexacosan-1-aminium 4-oxide (763 Da, Ligand)
- Membrane protein m (8 kDa, Protein from Usutu virus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Envelope protein e (53 kDa, Protein from Usutu virus)
- Trimethyl-[2-[[(2s,3s)-2-(octadecanoylamino)-3-oxidanyl-butoxy]-oxidanyl-phosphoryl]oxyethyl]azanium (537 Da, Ligand)
- Usutu virus (Virus from Usutu virus)