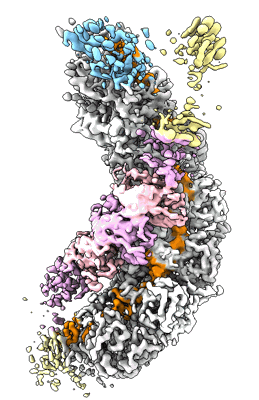
Cryo-EM structure of the prokaryotic SPARSA system complex
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8jkz
Q-score: 0.475
Zhen X, Xu X, Ye L, Xie S, Huang Z, Yang S, Wang Y, Li J, Long F, Ouyang S
Nat Commun (2024) 15 pp. 450-450 [ Pubmed: 38200015 DOI: doi:10.1038/s41467-023-44660-7 ]
- Ago (Complex)
- Sir2 superfamily protein (66 kDa, Protein from Geobacter sulfurreducens)
- Sir2 (Complex from Geobacter sulfurreducens PCA)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Piwi domain protein (53 kDa, Protein from Geobacter sulfurreducens)
- Sir2/ago (Complex from Geobacter sulfurreducens PCA)
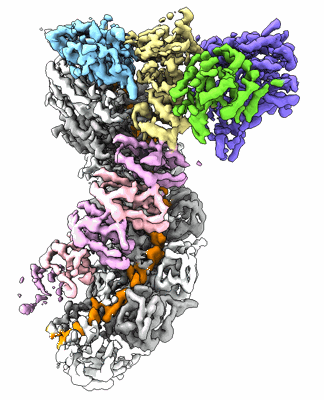
Cryo-EM structure of the prokaryotic SPARSA system complex
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8jl0
Q-score: 0.547
Zhen X, Xu X, Ye L, Xie S, Huang Z, Yang S, Wang Y, Li J, Long F, Ouyang S
Nat Commun (2024) 15 pp. 450-450 [ DOI: doi:10.1038/s41467-023-44660-7 Pubmed: 38200015 ]
- Sir2 (Complex from Geobacter sulfurreducens PCA)
- Dna (5'-d(p*ap*cp*gp*ap*cp*gp*tp*cp*tp*ap*ap*gp*ap*ap*ap*cp*cp*ap*tp*tp*ap*t)-3') (6 kDa, DNA from synthetic construct)
- Piwi domain protein (53 kDa, Protein from Geobacter sulfurreducens)
- Sir2 superfamily protein (66 kDa, Protein from Geobacter sulfurreducens)
- Ago (Complex)
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Dna/guide rna duplex (Complex)
- The sir2/ago/guide rna/recognition dna complex (Complex from Geobacter sulfurreducens PCA)
- Rna (5'-r(p*ap*up*ap*ap*up*gp*gp*up*up*up*cp*up*up*ap*gp*ap*cp*gp*up*cp*gp*u)-3') (6 kDa, RNA from synthetic construct)
Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8in8
Q-score: 0.504
Wang X, Li X, Yu G, Zhang L, Zhang C, Wang Y, Liao F, Wen Y, Yin H, Liu X, Wei Y, Li Z, Deng Z, Zhang H
Cell Res (2023) 33 pp. 699-711 [ DOI: doi:10.1038/s41422-023-00839-7 Pubmed: 37311833 ]
- Rna (5'-r(p*ap*up*ap*ap*up*gp*gp*up*up*up*cp*up*up*ap*gp*ap*cp*gp*up*cp*gp*up*up*u)-3') (7 kDa, RNA from Geobacter sulfurreducens (strain ATCC 51573 / DSM 12127 / PCA))
- Cryo-em structure of the target ssdna-bound sir2-apaz/ago-grna quaternary complex (Complex from Geobacter sulfurreducens (strain ATCC 51573 / DSM 12127 / PCA))
- Piwi domain protein (53 kDa, Protein from Geobacter sulfurreducens (strain ATCC 51573 / DSM 12127 / PCA))
- Magnesium ion (24 Da, Ligand)
- Sir2 superfamily protein (66 kDa, Protein from Geobacter sulfurreducens (strain ATCC 51573 / DSM 12127 / PCA))
- Dna (5'-d(p*ap*ap*cp*gp*ap*cp*gp*tp*cp*tp*ap*ap*gp*ap*ap*ap*cp*cp*ap*tp*tp*ap*a)-3') (7 kDa, DNA from Geobacter sulfurreducens (strain ATCC 51573 / DSM 12127 / PCA))
Intracytoplasmic membranes in Geobacter sulfurreducens
Howley E, Mangus A, Williams D, Torres CI
NPJ Biofilms Microbiomes (2023) 9 pp. 18-18 [ DOI: doi:10.1038/s41522-023-00384-6 Pubmed: 37029136 ]
Intracytoplasmic membranes in a tomogram of Geobacter sulfurreducens
Howley E, Mangus A, Williams D, Torres CI
NPJ Biofilms Microbiomes (2023) 9 pp. 18-18 [ DOI: doi:10.1038/s41522-023-00384-6 Pubmed: 37029136 ]
Electron cryotomogram of Geobacter sulfurreducens cell.
Howley E, Mangus A, Williams D, Torres CI
NPJ Biofilms Microbiomes (2023) 9 pp. 18-18 [ DOI: doi:10.1038/s41522-023-00384-6 Pubmed: 37029136 ]
Geobacter sulfurreducens whole cell tomogram with membrane invagination
Howley E, Mangus A, Williams D, Torres CI
NPJ Biofilms Microbiomes (2023) 9 pp. 18-18 [ DOI: doi:10.1038/s41522-023-00384-6 Pubmed: 37029136 ]
Cryo-EM structure of OmcZ nanowire from Geobacter sulfurreducens
Resolution: 3.4 Å
EM Method: Helical reconstruction
Fitted PDBs: 7lq5
Q-score: 0.462
Gu Y, Guberman-Pfeffer MJ, Srikanth V, Shen C, Giska F, Gupta K, Londer Y, Samatey FA, Batista VS, Malvankar NS
Nat Microbiol (2023) 8 pp. 284-298 [ Pubmed: 36732469 DOI: doi:10.1038/s41564-022-01315-5 ]
- Polymerized cytochrome omcz from geobacter sulfurreducens (Complex from Geobacter sulfurreducens)
- Heme c (618 Da, Ligand)
- Cytochrome c (27 kDa, Protein from Geobacter sulfurreducens PCA)
Cascade complex from type I-A CRISPR-Cas system
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7tr6
Q-score: 0.263
Hu C, Ni D, Nam KH, Majumdar S, McLean J, Stahlberg H, Terns MP, Ke A
Mol Cell (2022) 82 pp. 2754-2768.e5 [ DOI: doi:10.1016/j.molcel.2022.06.007 Pubmed: 35835111 ]
- Cas7a (36 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Crrna (45-mer) (14 kDa, RNA from Escherichia coli)
- Cas5a (29 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Cascade complex from pyrococcus furiosus type i-a crispr/cas system (350 kDa, Complex from Geobacter sulfurreducens)
- Cas11a (12 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Cas8a (38 kDa, Protein from Pyrococcus furiosus DSM 3638)
Cascade complex from type I-A CRISPR-Cas system
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7tr8
Q-score: 0.122
Hu C, Ni D, Nam KH, Majumdar S, McLean J, Stahlberg H, Terns MP, Ke A
Mol Cell (2022) 82 pp. 2754-2768.e5 [ DOI: doi:10.1016/j.molcel.2022.06.007 Pubmed: 35835111 ]
- Cas7a (36 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Nickel (ii) ion (58 Da, Ligand)
- Crrna (45-mer) (14 kDa, RNA from Escherichia coli)
- Cas5a (29 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Cas3 bound cascade complex from pyrococcus furiosus type i-a crispr/cas system (400 kDa, Complex from Geobacter sulfurreducens)
- Cas8 (38 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Crispr-associated endonuclease cas3-hd (26 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Cas11a (12 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Crispr-associated helicase cas3 (61 kDa, Protein from Pyrococcus furiosus DSM 3638)