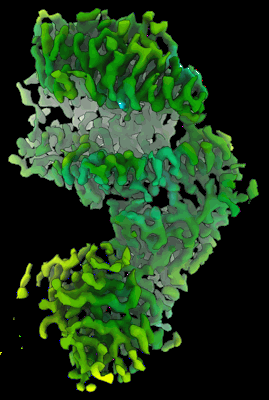
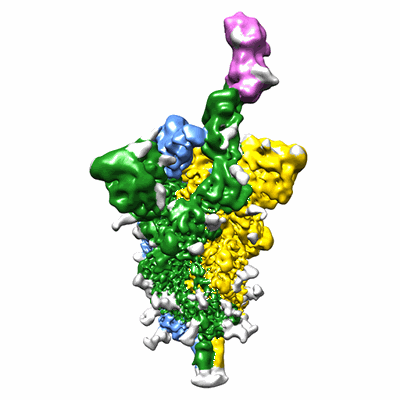
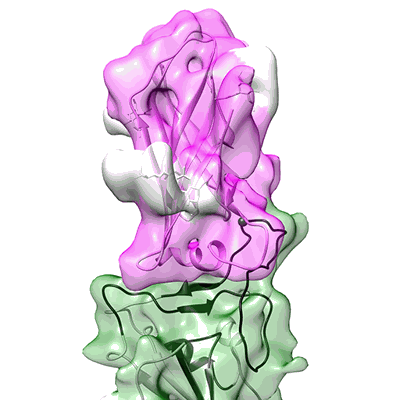
CryoEM structure of compound HNC-1664 bound with RdRP-RNA complex of SARS-CoV-2
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 8xko
Q-score: 0.486
Jia X, Jing X, Li M, Gao M, Zhong Y, Li E, Liu Y, Li R, Yao G, Liu Q, Zhou M, Hou Y, An L, Hong Y, Li S, Zhang J, Wang W, Zhang K, Gong P, Chiu S
Nat Commun (2024) 15 pp. 10750-10750 [ DOI: doi:10.1038/s41467-024-54918-3 Pubmed: 39737930 ]
- Rna (5'-r(p*up*up*cp*ap*up*gp*cp*up*ap*cp*gp*cp*gp*up*ap*g)-3') (5 kDa, RNA from synthetic construct)
- Cryoem structure of compound hnc-1644 bound with rdrp-rna (150 kDa, Complex from Severe acute respiratory syndrome coronavirus)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- [[(2~{r},3~{r},4~{s},5~{r})-4-fluoranyl-5-(5-iodanyl-4-methyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate (633 Da, Ligand)
- Rna (5'-r(p*cp*up*ap*cp*gp*cp*gp*up*ap*gp*cp*ap*up*g)-3') (4 kDa, RNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Non-structural protein 8 (23 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Rna-directed rna polymerase nsp12 (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8ekd
Q-score: 0.433
Desautels TA, Arrildt KT, Zemla AT, Lau EY, Zhu F, Ricci D, Cronin S, Zost SJ, Binshtein E, Scheaffer SM, Dadonaite B, Petersen BK, Engdahl TB, Chen E, Handal LS, Hall L, Goforth JW, Vashchenko D, Nguyen S, Weilhammer DR, Lo JK, Rubinfeld B, Saada EA, Weisenberger T, Lee TH, Whitener B, Case JB, Ladd A, Silva MS, Haluska RM, Grzesiak EA, Earnhart CG, Hopkins S, Bates TW, Thackray LB, Segelke BW, Lillo AM, Sundaram S, Bloom JD, Diamond MS, Crowe Jr JE, Carnahan RH, Faissol DM
Nature (2024) 629 pp. 878-885 [ Pubmed: 38720086 DOI: doi:10.1038/s41586-024-07385-1 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Fab llnl-199 lc (12 kDa, Protein from Homo sapiens)
- Sars cov2 omicron ba.2 rbd complex with fab llnl-199 (Complex)
- Sars cov2 omicron ba.2 rbd (Complex from Severe acute respiratory syndrome coronavirus)
- Fab llnl-199 hc (14 kDa, Protein from Homo sapiens)
- Spike protein s2' (20 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Fab llnl-199 (Complex from Homo sapiens)
SARS-CoV-2 Omicron 1-RBD up Spike trimer complexed with three XG005 molecules
Resolution: 3.74 Å
EM Method: Single-particle
Fitted PDBs: 7ycy
Q-score: 0.26
To Be Published
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Xg005-vh (13 kDa, Protein from Homo sapiens)
- Omicron rbd with xg005 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Xg005-vl (11 kDa, Protein from Homo sapiens)
SARS-CoV-2 Omicron 1-RBD up spike trimer complexed with two XG005 Fab
Resolution: 3.58 Å
EM Method: Single-particle
Fitted PDBs: 7yd0
Q-score: 0.267
To Be Published
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Xg005-vl (21 kDa, Protein from Homo sapiens)
- Omicron rbd with xg005 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Xg005-vh (23 kDa, Protein from Homo sapiens)
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8fxb
Q-score: 0.469
Addetia A, Piccoli L, Case JB, Park YJ, Beltramello M, Guarino B, Dang H, de Melo GD, Pinto D, Sprouse K, Scheaffer SM, Bassi J, Silacci-Fregni C, Muoio F, Dini M, Vincenzetti L, Acosta R, Johnson D, Subramanian S, Saliba C, Giurdanella M, Lombardo G, Leoni G, Culap K, McAlister C, Rajesh A, Dellota Jr E, Zhou J, Farhat N, Bohan D, Noack J, Chen A, Lempp FA, Quispe J, Kergoat L, Larrous F, Cameroni E, Whitener B, Giannini O, Cippa P, Ceschi A, Ferrari P, Franzetti-Pellanda A, Biggiogero M, Garzoni C, Zappi S, Bernasconi L, Kim MJ, Rosen LE, Schnell G, Czudnochowski N, Benigni F, Franko N, Logue JK, Yoshiyama C, Stewart C, Chu H, Bourhy H, Schmid MA, Purcell LA, Snell G, Lanzavecchia A, Diamond MS, Corti D, Veesler D
Nature (2023) 621 pp. 592-601 [ DOI: doi:10.1038/s41586-023-06487-6 Pubmed: 37648855 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 xbb.1 spike rbd bound to the human ace2 ectodomain and the s309 neutralizing antibody fab fragment (Complex from Severe acute respiratory syndrome coronavirus 2)
- S309 heavy chain (50 kDa, Protein from Homo sapiens)
- Processed angiotensin-converting enzyme 2 (74 kDa, Protein from Homo sapiens)
- Sars-cov-2 xbb.1 spike rbd bound to the human ace2 ectodomain and the s309 neutralizing antibody fab fragment (Complex from Severe acute respiratory syndrome coronavirus 2)
- S309 light chain (23 kDa, Protein from Homo sapiens)
- Spike protein s1 (28 kDa, Protein from Severe acute respiratory syndrome coronavirus)
SARS Cov2 Spike RBD in complex with Fab47
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8a95
Q-score: 0.269
Pushparaj P, Nicoletto A, Sheward DJ, Das H, Castro Dopico X, Perez Vidakovics L, Hanke L, Chernyshev M, Narang S, Kim S, Fischbach J, Ekstrom S, McInerney G, Hallberg BM, Murrell B, Corcoran M, Karlsson Hedestam GB
Immunity (2023) 56 pp. 193-206.e7 [ Pubmed: 36574772 DOI: doi:10.1016/j.immuni.2022.12.005 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov2 trimeric spike in 1-up conformation in complex with three fab47 (142 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Fab47 (light chain variable domain) (11 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike in complex with fab47 (629 MDa, Complex from Homo sapiens)
- Fab47 (heavy chain variable domain) (13 kDa, Protein from Homo sapiens)
Cryo-EM structure of the SARS-CoV-2 Spike bound to TMEM106B
Baggen J, Jacquemyn M, Persoons L, Vanstreels E, Pye VE, Wrobel AG, Calvaresi V, Martin SR, Roustan C, Cronin NB, Reading E, Thibaut HJ, Vercruysse T, Maes P, De Smet F, Yee A, Nivitchanyong T, Roell M, Franco-Hernandez N, Rhinn H, Mamchak AA, Ah Young-Chapon M, Brown E, Cherepanov P, Daelemans D
Cell (2023) 186 pp. 3427-3442.e22 [ DOI: doi:10.1016/j.cell.2023.06.005 Pubmed: 37421949 ]
- Sars-cov-2 spike ectodomain, hexa-pro stabilized; belgium/ghb-03021 variant (asp-484) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike ectodomain, hexa-pro stabilized; belgium/ghb-03021 variant (asp-484) (Protein from Severe acute respiratory syndrome coronavirus 2)
- Tmem106b lumenal domain (Protein from Homo sapiens)
- Tmem106b lumenal domain (Complex from Homo sapiens)
- Trimeric sars-cov-2 spike bound to one molecule of tmem106b (450 kDa, Complex)
Local refinement of the SARS-CoV-2 Spike RBD bound to TMEM106B
Baggen J, Jacquemyn M, Persoons L, Vanstreels E, Pye VE, Wrobel AG, Calvaresi V, Martin SR, Roustan C, Cronin NB, Reading E, Thibaut HJ, Vercruysse T, Maes P, De Smet F, Yee A, Nivitchanyong T, Roell M, Franco-Hernandez N, Rhinn H, Mamchak AA, Ah Young-Chapon M, Brown E, Cherepanov P, Daelemans D
Cell (2023) 186 pp. 3427-3442.e22 [ DOI: doi:10.1016/j.cell.2023.06.005 Pubmed: 37421949 ]
- Sars-cov-2 spike ectodomain (belgium/ghb-03021) stabilized in trimeric pre-fusion state (Complex from Severe acute respiratory syndrome coronavirus 2)
- Tmem106b lumenall domain (Protein from Homo sapiens)
- Tmem106b lumenall domain (Complex from Homo sapiens)
- Trimeric sars-cov-2 spike bound to one molecule of tmem106b (450 kDa, Complex)
- Sars-cov-2 spike ectodomain (belgium/ghb-03021) stabilized in trimeric pre-fusion state (Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8fdw
Q-score: 0.327
Shi W, Cai Y, Zhu H, Peng H, Voyer J, Rits-Volloch S, Cao H, Mayer ML, Song K, Xu C, Lu J, Zhang J, Chen B
Nature (2023) 619 pp. 403-409 [ DOI: doi:10.1038/s41586-023-06273-4 Pubmed: 37285872 ]
- Alpha-d-mannopyranose (180 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike protein s2 (68 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Sras-cov-2 postfusion spike protein in nanodisc (210 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
SARS CoV2 Spike in the 2-up state in complex with Fab47
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 8a94
Q-score: 0.303
Pushparaj P, Nicoletto A, Sheward DJ, Das H, Castro Dopico X, Perez Vidakovics L, Hanke L, Chernyshev M, Narang S, Kim S, Fischbach J, Ekstrom S, McInerney G, Hallberg BM, Murrell B, Corcoran M, Karlsson Hedestam GB
Immunity (2023) 56 pp. 193-206.e7 [ Pubmed: 36574772 DOI: doi:10.1016/j.immuni.2022.12.005 ]
- Fab47 (heavy chain variable domain) (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike in complex with fab47 (629 MDa, Complex from Homo sapiens)
- Fab47 (light chain variable domain) (11 kDa, Protein from Homo sapiens)
- Sars-cov2 spike in 2-up conformation in complex with fab47 (142 kDa, Protein from Severe acute respiratory syndrome coronavirus)