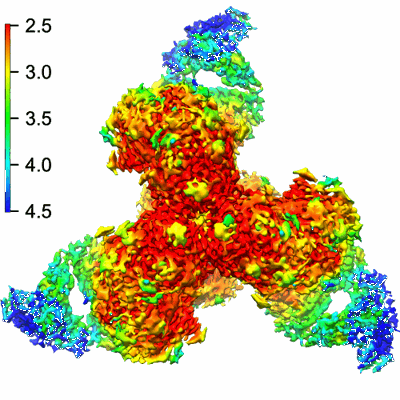
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8euu
Q-score: 0.545
Banach BB, Pletnev S, Olia AS, Xu K, Zhang B, Rawi R, Bylund T, Doria-Rose NA, Nguyen TD, Fahad AS, Lee M, Lin BC, Liu T, Louder MK, Madan B, McKee K, O'Dell S, Sastry M, Schon A, Bui N, Shen CH, Wolfe JR, Chuang GY, Mascola JR, Kwong PD, DeKosky BJ
Nat Commun (2023) 14 pp. 7593-7593 [ DOI: doi:10.1038/s41467-023-42098-5 Pubmed: 37989731 ]
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Vrc34.01 fab variable heavy chain (23 kDa, Protein from Homo sapiens)
- Envelope glycoprotein gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Vrc34.01 fab variable light chain (23 kDa, Protein from Homo sapiens)
- Bg505 ds-sosip vrc34.01 fab complex (Complex from Human immunodeficiency virus 1)
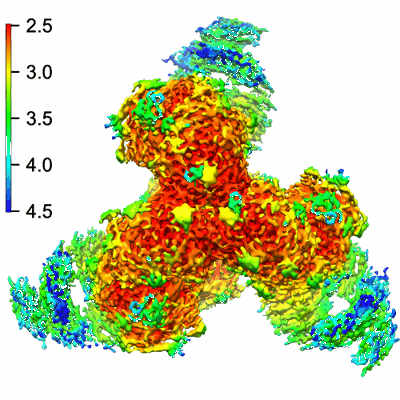
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8euv
Q-score: 0.586
Banach BB, Pletnev S, Olia AS, Xu K, Zhang B, Rawi R, Bylund T, Doria-Rose NA, Nguyen TD, Fahad AS, Lee M, Lin BC, Liu T, Louder MK, Madan B, McKee K, O'Dell S, Sastry M, Schon A, Bui N, Shen CH, Wolfe JR, Chuang GY, Mascola JR, Kwong PD, DeKosky BJ
Nat Commun (2023) 14 pp. 7593-7593 [ DOI: doi:10.1038/s41467-023-42098-5 Pubmed: 37989731 ]
- Water (18 Da, Ligand)
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Envelope glycoprotein gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Vrc34.01-combo1 fab variable light chain (23 kDa, Protein from Homo sapiens)
- Bg505 ds-sosip vrc34.01-combo1 fab complex (Complex from Human immunodeficiency virus 1)
- Vrc34.01-combo1 fab variable heavy chain (23 kDa, Protein from Homo sapiens)
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8euw
Q-score: 0.553
Banach BB, Pletnev S, Olia AS, Xu K, Zhang B, Rawi R, Bylund T, Doria-Rose NA, Nguyen TD, Fahad AS, Lee M, Lin BC, Liu T, Louder MK, Madan B, McKee K, O'Dell S, Sastry M, Schon A, Bui N, Shen CH, Wolfe JR, Chuang GY, Mascola JR, Kwong PD, DeKosky BJ
Nat Commun (2023) 14 pp. 7593-7593 [ DOI: doi:10.1038/s41467-023-42098-5 Pubmed: 37989731 ]
- Vrc34.01-mm28 fab variable light chain (23 kDa, Protein from Homo sapiens)
- Vrc34.01-mm28 fab variable heavy chain (23 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Envelope glycoprotein gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Bg505 ds-sosip vrc34.01-mm28 fab complex (Complex from Human immunodeficiency virus 1)
Cryo-EM structure of prefusion SARS-CoV-2 spike glycoprotein in complex with 910-30 Fab
Resolution: 4.75 Å
EM Method: Single-particle
Fitted PDBs: 7ks9
Q-score: 0.223
Banach BB, Cerutti G, Fahad AS, Shen CH, Oliveira De Souza M, Katsamba PS, Tsybovsky Y, Wang P, Nair MS, Huang Y, Francino-Urdaniz IM, Steiner PJ, Gutierrez-Gonzalez M, Liu L, Lopez Acevedo SN, Nazzari AF, Wolfe JR, Luo Y, Olia AS, Teng IT, Yu J, Zhou T, Reddem ER, Bimela J, Pan X, Madan B, Laflin AD, Nimrania R, Yuen KY, Whitehead TA, Ho DD, Kwong PD, Shapiro L, DeKosky BJ
Cell Rep (2021) 37 pp. 109771-109771 [ DOI: doi:10.1016/j.celrep.2021.109771 Pubmed: 34587480 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Prefusion sars-cov-2 spike glycoprotein in complex with 910-30 fab (Complex from Homo sapiens)
- 910-30 fab light chain (23 kDa, Protein from Homo sapiens)
- 910-30 fab heavy chain (23 kDa, Protein from Homo sapiens)
Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down)
Resolution: 3.45 Å
EM Method: Single-particle
Fitted PDBs: 6xf5
Q-score: 0.365
Zhou T, Teng IT, Olia AS, Cerutti G, Gorman J, Nazzari A, Shi W, Tsybovsky Y, Wang L, Wang S, Zhang B, Zhang Y, Katsamba PS, Petrova Y, Banach BB, Fahad AS, Liu L, Acevedo SNL, Madan B, de Souza MO, Pan X, Wang P, Wolfe JR, Yin M, Ho DD, Phung E, DiPiazza A, Chang L, Abiona O, Corbett KS, DeKosky BJ, Graham BS, Mascola JR, Misasi J, Ruckwardt T, Sullivan NJ, Shapiro L, Kwong PD
SSRN (2020) pp. 3639618-3639618 [ Pubmed: 32742241 DOI: doi:10.2139/ssrn.3639618 ]
Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up)
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6xf6
Q-score: 0.295
Zhou T, Teng IT, Olia AS, Cerutti G, Gorman J, Nazzari A, Shi W, Tsybovsky Y, Wang L, Wang S, Zhang B, Zhang Y, Katsamba PS, Petrova Y, Banach BB, Fahad AS, Liu L, Acevedo SNL, Madan B, de Souza MO, Pan X, Wang P, Wolfe JR, Yin M, Ho DD, Phung E, DiPiazza A, Chang L, Abiona O, Corbett KS, DeKosky BJ, Graham BS, Mascola JR, Misasi J, Ruckwardt T, Sullivan NJ, Shapiro L, Kwong PD
SSRN (2020) pp. 3639618-3639618 [ Pubmed: 32742241 DOI: doi:10.2139/ssrn.3639618 ]