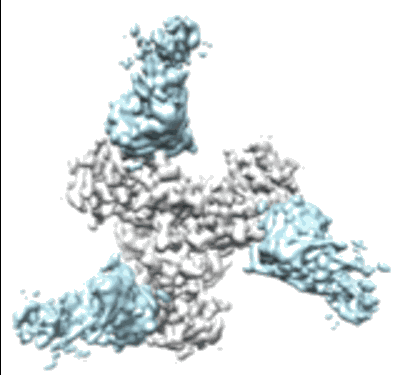
Resolution: 4.32 Å
EM Method: Single-particle
Fitted PDBs: 8dy6
Q-score: 0.355
Wiehe K, Saunders KO, Stalls V, Cain DW, Venkatayogi S, Martin Beem JS, Berry M, Evangelous T, Henderson R, Hora B, Xia SM, Jiang C, Newman A, Bowman C, Lu X, Bryan ME, Bal J, Sanzone A, Chen H, Eaton A, Tomai MA, Fox CB, Tam YK, Barbosa C, Bonsignori M, Muramatsu H, Alam SM, Montefiori DC, Williams WB, Pardi N, Tian M, Weissman D, Alt FW, Acharya P, Haynes BF
Cell Host Microbe (2024) 32 pp. 693-709.e7 [ DOI: doi:10.1016/j.chom.2024.04.006 Pubmed: 38670093 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Mu89+s27y light chain (11 kDa, Protein from Mus musculus)
- V3-glycan vaccine induced antibody mu89+s27y in complex with ch848.d949.10.17_n133d_n138t.ds.sosip.664 hiv env trimer (360 kDa, Complex from Human immunodeficiency virus 1)
- Envelope glycoprotein gp160 (72 kDa, Protein from Human immunodeficiency virus 1)
- Mu89+s27y heavy chain (16 kDa, Protein from Mus musculus)
Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 7lua
Q-score: 0.267
Williams WB, Meyerhoff RR, Edwards RJ, Li H, Manne K, Nicely NI, Henderson R, Zhou Y, Janowska K, Mansouri K, Gobeil S, Evangelous T, Hora B, Berry M, Abuahmad AY, Sprenz J, Deyton M, Stalls V, Kopp M, Hsu AL, Borgnia MJ, Stewart-Jones GBE, Lee MS, Bronkema N, Moody MA, Wiehe K, Bradley T, Alam SM, Parks RJ, Foulger A, Oguin T, Sempowski GD, Bonsignori M, LaBranche CC, Montefiori DC, Seaman M, Santra S, Perfect J, Francica JR, Lynn GM, Aussedat B, Walkowicz WE, Laga R, Kelsoe G, Saunders KO, Fera D, Kwong PD, Seder RA, Bartesaghi A, Shaw GM, Acharya P, Haynes BF
Cell (2021) 184 pp. 2955- [ Pubmed: 34019795 DOI: doi:10.1016/j.cell.2021.04.042 ]
- Ch848 sosip gp120 (52 kDa, Protein from Human immunodeficiency virus 1)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Env polyprotein (16 kDa, Protein from Simian-Human immunodeficiency virus)
- Cryo-em structure of dh898.1 fab-dimer bound near the cd4 binding site of hiv-1 env ch848 sosip trimer. (Complex from Human immunodeficiency virus 1)
- Dh898.1 heavy chain (23 kDa, Protein from Homo sapiens)
- Dh898.1 light chain (23 kDa, Protein from Homo sapiens)
Williams WB, Meyerhoff RR, Edwards RJ, Li H, Nicely NI, Henderson R, Zhou Y, Janowska K, Mansouri K, Manne K, Stalls V, Hsu AL, Borgnia MJ, Stewart-Jones G, Lee MS, Bronkema N, Perfect J, Moody MA, Wiehe K, Bradley T, Kepler TB, Alam SM, Foulger A, Bonsignori M, LaBranche CC, Montefiori DC, Seaman M, Santra S, Francica JR, Lynn GM, Aussedat B, Walkowicz WE, Laga R, Kelso G, Saunders KO, Fera D, Kwong PD, Seder RA, Bartesaghi A, Shaw GM, Acharya P, Haynes BF
bioRxiv (2020) https://doi.org/10.1101/2020.06.30.178921 [ DOI: doi:10.1101/2020.06.30.178921 ]
- Antibody dh898.1 fragment antigen binding (fab) dimer (Complex from Macaca mulatta)
- Hiv env ch848.3.d0949.10.17chim.6r.ds.sosip.664 (Protein from Human immunodeficiency virus 1)
- Antibody dh898.1 fragment antigen binding (fab) heavy chain (Protein from Macaca mulatta)
- Hiv env ch848.3.d0949.10.17chim.6r.ds.sosip.664 trimer (Complex from Human immunodeficiency virus 1)
- Antibody dh898.1 fab-dimer bound near the cd4 binding site of hiv env ch848.3.d0949.10.17chim.6r.ds.sosip.664 trimer (94 kDa, Complex)
- Antibody dh898.1 fragment antigen binding (fab) light chain (Protein from Macaca mulatta)
Cryo-electron microscopy reconstruction of locally refined antibody DH898.1 Fab-dimer
Resolution: 4.8 Å
EM Method: Single-particle
Fitted PDBs: 7l6m
Q-score: 0.267
Williams WB, Meyerhoff RR, Edwards RJ, Li H, Manne K, Nicely NI, Henderson R, Zhou Y, Janowska K, Mansouri K, Gobeil S, Evangelous T, Hora B, Berry M, Abuahmad AY, Sprenz J, Deyton M, Stalls V, Kopp M, Hsu AL, Borgnia MJ, Stewart-Jones GBE, Lee MS, Bronkema N, Moody MA, Wiehe K, Bradley T, Alam SM, Parks RJ, Foulger A, Oguin T, Sempowski GD, Bonsignori M, LaBranche CC, Montefiori DC, Seaman M, Santra S, Perfect J, Francica JR, Lynn GM, Aussedat B, Walkowicz WE, Laga R, Kelsoe G, Saunders KO, Fera D, Kwong PD, Seder RA, Bartesaghi A, Shaw GM, Acharya P, Haynes BF
Cell (2021) 184 pp. 2955-2972.e25 [ Pubmed: 34019795 DOI: doi:10.1016/j.cell.2021.04.042 ]
Cryo-EM structure of HIV-1 neutralizing antibody DH270 UCA3 in complex with CH848 10.17DT Env
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 6um5
Q-score: 0.347
Saunders KO, Wiehe K, Tian M, Acharya P, Bradley T, Alam SM, Go EP, Scearce R, Sutherland L, Henderson R, Hsu AL, Borgnia MJ, Chen H, Lu X, Wu NR, Watts B, Jiang C, Easterhoff D, Cheng HL, McGovern K, Waddicor P, Chapdelaine-Williams A, Eaton A, Zhang J, Rountree W, Verkoczy L, Tomai M, Lewis MG, Desaire HR, Edwards RJ, Cain DW, Bonsignori M, Montefiori D, Alt FW, Haynes BF
Science (2019) 366 [ DOI: doi:10.1126/science.aay7199 Pubmed: 31806786 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Dh270 uca3 fab bound to hiv-1 env ch848 10.17dt (360 kDa, Complex from Human immunodeficiency virus 1)
- Envelope glycoprotein gp160 (17 kDa, Protein from Human immunodeficiency virus 1)
- Ch848 10.17 dt gp120 (51 kDa, Protein from Human immunodeficiency virus 1)
- Dh270 uca3 fab light chain (22 kDa, Protein from Homo sapiens)
- Dh270 uca3 fab heavy chain (25 kDa, Protein from Homo sapiens)
Cryo-EM structure of HIV-1 neutralizing antibody DH270.6 in complex with CH848 10.17DT Env
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 6um6
Q-score: 0.318
Saunders KO, Wiehe K, Tian M, Acharya P, Bradley T, Alam SM, Go EP, Scearce R, Sutherland L, Henderson R, Hsu AL, Borgnia MJ, Chen H, Lu X, Wu NR, Watts B, Jiang C, Easterhoff D, Cheng HL, McGovern K, Waddicor P, Chapdelaine-Williams A, Eaton A, Zhang J, Rountree W, Verkoczy L, Tomai M, Lewis MG, Desaire HR, Edwards RJ, Cain DW, Bonsignori M, Montefiori D, Alt FW, Haynes BF
Science (2019) 366 [ DOI: doi:10.1126/science.aay7199 Pubmed: 31806786 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Dh270.6 heavy chain (25 kDa, Protein from Homo sapiens)
- Complex of antibody dh270.6 with hiv-1 env ch848 10.17dt (360 kDa, Complex from Human immunodeficiency virus 1)
- Ch848 10.17dt gp120 (51 kDa, Protein from Human immunodeficiency virus 1)
- Ch848 10.17dt gp41 (18 kDa, Protein from Human immunodeficiency virus 1)
- Dh270.6 light chain (22 kDa, Protein from Homo sapiens)
Cryo-EM structure of CH235UCA bound to Man5-enriched CH505.N279K.G458Y.SOSIP.664
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 6uda
Q-score: 0.325
LaBranche CC, Henderson R, Hsu A, Behrens S, Chen X, Zhou T, Wiehe K, Saunders KO, Alam SM, Bonsignori M, Borgnia MJ, Sattentau QJ, Eaton A, Greene K, Gao H, Liao HX, Williams WB, Peacock J, Tang H, Perez LG, Edwards RJ, Kepler TB, Korber BT, Kwong PD, Mascola JR, Acharya P, Haynes BF, Montefiori DC
PLoS Pathog (2019) 15 pp. e1008026-e1008026 [ DOI: doi:10.1371/journal.ppat.1008026 Pubmed: 31527908 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ch505.n279k.g458y.sosip.664 gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
- Ch235 uca heavy chain fab (26 kDa, Protein from Homo sapiens)
- Ch235 uca light chain fab (25 kDa, Protein from Homo sapiens)
- Complex of the ch235 unmutated common ancestor (uca) bound to man5-enriched ch505.n279k.g458y.sosip.664 (360 kDa, Complex from Human immunodeficiency virus 1)
- Ch505.n279k.g458y.sosip.664 gp41 (18 kDa, Protein from Human immunodeficiency virus 1)