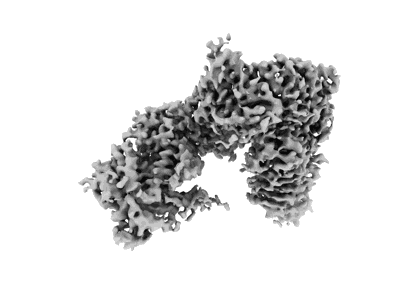
X. laevis CMG dimer bound to dimeric DONSON - without ATPase
Resolution: 3.14 Å
EM Method: Single-particle
Fitted PDBs: 8q6o
Q-score: 0.471
Cvetkovic MA, Passaretti P, Butryn A, Reynolds-Winczura A, Kingsley G, Skagia A, Fernandez-Cuesta C, Poovathumkadavil D, George R, Chauhan AS, Jhujh SS, Stewart GS, Gambus A, Costa A
Mol Cell (2023) 83 pp. 4017- [ Pubmed: 37820732 DOI: doi:10.1016/j.molcel.2023.09.029 ]
- Zinc ion (65 Da, Ligand)
- Dna replication complex gins protein psf1 (23 kDa, Protein from Xenopus laevis)
- Dna replication licensing factor mcm5-a (82 kDa, Protein from Xenopus laevis)
- Dna replication licensing factor mcm2 (100 kDa, Protein from Xenopus laevis)
- Dna replication licensing factor mcm4-b (97 kDa, Protein from Xenopus laevis)
- Maternal dna replication licensing factor mcm6 (92 kDa, Protein from Xenopus laevis)
- Dna replication complex gins protein psf3 (23 kDa, Protein from Xenopus laevis)
- Dna replication complex gins protein psf2 (21 kDa, Protein from Xenopus laevis)
- Maternal dna replication licensing factor mcm3 (90 kDa, Protein from Xenopus laevis)
- Protein downstream neighbor of son homolog (63 kDa, Protein from Xenopus laevis)
- X. laevis cmg dimer bound to dimeric donson (1 MDa, Complex from Xenopus laevis)
- Dna replication licensing factor mcm7-b (81 kDa, Protein from Xenopus laevis)
- Cell division control protein 45 homolog (65 kDa, Protein from Xenopus laevis)
- Dna replication complex gins protein sld5 (25 kDa, Protein from Xenopus laevis)
X. laevis CMG dimer bound to dimeric DONSON - MCM ATPase
Resolution: 3.53 Å
EM Method: Single-particle
Fitted PDBs: 8q6p
Q-score: 0.369
Cvetkovic MA, Passaretti P, Butryn A, Reynolds-Winczura A, Kingsley G, Skagia A, Fernandez-Cuesta C, Poovathumkadavil D, George R, Chauhan AS, Jhujh SS, Stewart GS, Gambus A, Costa A
Mol Cell (2023) 83 pp. 4017- [ Pubmed: 37820732 DOI: doi:10.1016/j.molcel.2023.09.029 ]
- Dna replication licensing factor mcm5-a (82 kDa, Protein from Xenopus laevis)
- Dna replication licensing factor mcm2 (100 kDa, Protein from Xenopus laevis)
- Dna replication licensing factor mcm4-b (97 kDa, Protein from Xenopus laevis)
- Maternal dna replication licensing factor mcm6 (92 kDa, Protein from Xenopus laevis)
- Maternal dna replication licensing factor mcm3 (90 kDa, Protein from Xenopus laevis)
- Protein downstream neighbor of son homolog (63 kDa, Protein from Xenopus laevis)
- X. laevis cmg dimer bound to dimeric donson (1 MDa, Complex from Xenopus laevis)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Dna replication licensing factor mcm7-b (81 kDa, Protein from Xenopus laevis)
Single CMG purified from replicating Xenopus egg extracts
Cvetkovic MA, Passaretti P, Butryn A, Reynolds-Winczura A, Kingsley G, Skagia A, Fernandez-Cuesta C, Poovathumkadavil D, George R, Chauhan AS, Jhujh SS, Stewart GS, Gambus A, Costa A
Mol Cell (2023) 83 pp. 4017-4031.e9 [ Pubmed: 37820732 DOI: doi:10.1016/j.molcel.2023.09.029 ]
- Dna replication licensing factor mcm4 - xenopus laevis (Protein from Xenopus laevis)
- Single cmg purified from replicating xenopus egg extracts (700 kDa, Complex from Xenopus laevis)
- Dna replication complex gins protein psf3 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication complex gins protein sld5 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication complex gins protein psf1 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication licensing factor mcm2 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication licensing factor mcm5 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication licensing factor mcm3 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication complex gins protein psf2 - xenopus laevis (Protein from Xenopus laevis)
- Cell division control protein 45 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication licensing factor mcm6 - xenopus laevis (Protein from Xenopus laevis)
- Dna replication licensing factor mcm7 - xenopus laevis (Protein from Xenopus laevis)
Mot1:TBP:DNA - pre-hydrolysis state
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7zke
Q-score: 0.377
Woike S, Eustermann S, Jung J, Wenzl SJ, Hagemann G, Bartho J, Lammens K, Butryn A, Herzog F, Hopfner KP
Nat Struct Mol Biol (2023) 30 pp. 640-649 [ DOI: doi:10.1038/s41594-023-00966-0 Pubmed: 37106137 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Mot1:tbp:dna complex in pre-hydrolysis conformation (Complex from Chaetomium thermophilum)
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Putative tata-box binding protein (29 kDa, Protein from Chaetomium thermophilum)
- Helicase-like protein (211 kDa, Protein from Chaetomium thermophilum)
- Magnesium ion (24 Da, Ligand)
- Beryllium trifluoride ion (66 Da, Ligand)
Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7zb5
Q-score: 0.456
Woike S, Eustermann S, Jung J, Wenzl SJ, Hagemann G, Bartho J, Lammens K, Butryn A, Herzog F, Hopfner KP
Nat Struct Mol Biol (2023) 30 pp. 640-649 [ DOI: doi:10.1038/s41594-023-00966-0 Pubmed: 37106137 ]
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Mot1(1-1836):tbp:dna complex dimer (Complex from Chaetomium thermophilum)
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Putative tata-box binding protein (29 kDa, Protein from Chaetomium thermophilum)
- Helicase-like protein (206 kDa, Protein from Chaetomium thermophilum)
Mot1E1434Q:TBP:DNA - substrate recognition state
Resolution: 5.1 Å
EM Method: Single-particle
Fitted PDBs: 7z7n
Q-score: 0.242
Woike S, Eustermann S, Jung J, Wenzl SJ, Hagemann G, Bartho J, Lammens K, Butryn A, Herzog F, Hopfner KP
Nat Struct Mol Biol (2023) 30 pp. 640-649 [ DOI: doi:10.1038/s41594-023-00966-0 Pubmed: 37106137 ]
- Mot1e1434q:tbp:dna complex (Complex from Chaetomium thermophilum)
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Putative tata-box binding protein (29 kDa, Protein from Chaetomium thermophilum)
- Helicase-like protein (211 kDa, Protein from Chaetomium thermophilum)
Woike S, Eustermann S, Jung J, Wenzl SJ, Hagemann G, Bartho J, Lammens K, Butryn A, Herzog F, Hopfner KP
Nat Struct Mol Biol (2023) 30 pp. 640-649 [ DOI: doi:10.1038/s41594-023-00966-0 Pubmed: 37106137 ]
Mot1:TBP:DNA - post hydrolysis state
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7z8s
Q-score: 0.403
Woike S, Eustermann S, Jung J, Wenzl SJ, Hagemann G, Bartho J, Lammens K, Butryn A, Herzog F, Hopfner KP
Nat Struct Mol Biol (2023) 30 pp. 640-649 [ DOI: doi:10.1038/s41594-023-00966-0 Pubmed: 37106137 ]
- Mot1:tbp:dna post hydrolysis complex (Complex from Chaetomium thermophilum)
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Dna (36-mer) (11 kDa, DNA from DNA molecule)
- Putative tata-box binding protein (29 kDa, Protein from Chaetomium thermophilum)
- Helicase-like protein (211 kDa, Protein from Chaetomium thermophilum)
Structure of Mot1 in complex with TBP, NC2 and DNA
Butryn A, Schuller JM, Stoehr G, Runge-Wollmann P, Foerster F, Auble DT, Hopfner K-P
eLife (2015) 4 pp. 403-407 [ Pubmed: 26258880 DOI: doi:10.7554/eLife.07432 ]