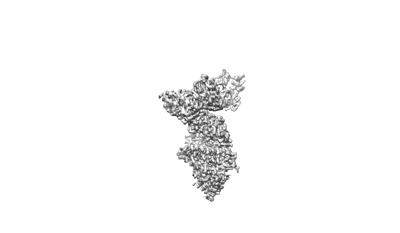
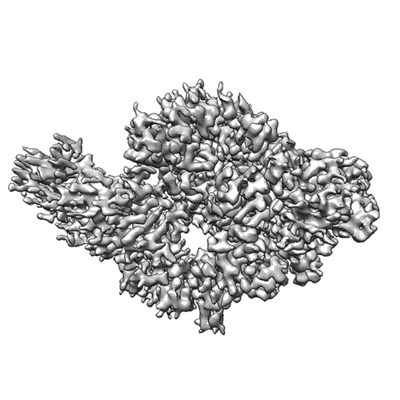
S-RBD (Omicron BA.2.75) in complex with PD of ACE2
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8i9f
Q-score: 0.573
Li Y, Ren C, Shen Y, Zhang Y, Chen J, Zheng J, Tian R, Cao L, Yan R
Cell Discov (2023) 9 pp. 108-108 [ DOI: doi:10.1038/s41421-023-00607-2 Pubmed: 37919275 ]
- S-rbd (omicron ba.2.75) (Complex from Homo sapiens)
- Spike protein s2' (25 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Processed angiotensin-converting enzyme 2 (71 kDa, Protein from Homo sapiens)
- S-rbd (omicron ba.2.75) in complex with pd of ace2 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Pd of ace2 (Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Top-down design of protein architectures with reinforcement learning
Resolution: 2.93 Å
EM Method: Single-particle
Fitted PDBs: 8f53
Q-score: 0.555
Lutz ID, Wang S, Norn C, Courbet A, Borst AJ, Zhao YT, Dosey A, Cao L, Xu J, Leaf EM, Treichel C, Litvicov P, Li Z, Goodson AD, Rivera-Sanchez P, Bratovianu AM, Baek M, King NP, Ruohola-Baker H, Baker D
Science (2023) 380 pp. 266-273 [ Pubmed: 37079676 DOI: doi:10.1126/science.adf6591 ]
- Rc_i_2 (Complex from Escherichia coli)
- Rc_i_2 (5 kDa, Protein from synthetic construct)
Top-down design of protein architectures with reinforcement learning
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8f54
Q-score: 0.61
Lutz ID, Wang S, Norn C, Courbet A, Borst AJ, Zhao YT, Dosey A, Cao L, Xu J, Leaf EM, Treichel C, Litvicov P, Li Z, Goodson AD, Rivera-Sanchez P, Bratovianu AM, Baek M, King NP, Ruohola-Baker H, Baker D
Science (2023) 380 pp. 266-273 [ Pubmed: 37079676 DOI: doi:10.1126/science.adf6591 ]
- Rc_i_1 (7 kDa, Protein from synthetic construct)
- Rc_i_1 (Complex from Escherichia coli)
Cryo-EM structure of PDF-2180 Spike glycoprotein
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7u6r
Q-score: 0.63
Xiong Q, Cao L, Ma C, Tortorici MA, Liu C, Si J, Liu P, Gu M, Walls AC, Wang C, Shi L, Tong F, Huang M, Li J, Zhao C, Shen C, Chen Y, Zhao H, Lan K, Corti D, Veesler D, Wang X, Yan H
Nature (2022) 612 pp. 748-757 [ DOI: doi:10.1038/s41586-022-05513-3 Pubmed: 36477529 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Predict/pdf-2180 (Virus from PREDICT/PDF-2180)
- Pdf-2180 spike glycoprotein (147 kDa, Protein from Bat coronavirus)
Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex
Resolution: 2.57 Å
EM Method: Single-particle
Fitted PDBs: 7xcr
Q-score: 0.571
Ai H, Sun M, Liu A, Sun Z, Liu T, Cao L, Liang L, Qu Q, Li Z, Deng Z, Tong Z, Chu G, Tian X, Deng H, Zhao S, Li JB, Lou Z, Liu L
Nat Chem Biol (2022) 18 pp. 972-980 [ Pubmed: 35739357 DOI: doi:10.1038/s41589-022-01067-7 ]
- Complex of dot1l(1-416) and h2bk34ub nucleosome (Complex from Homo sapiens)
- Dna (146-mer) (44 kDa, DNA from synthetic construct)
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Histone domain-containing protein (11 kDa, Protein from synthetic construct)
- Dna (146-mer) (45 kDa, DNA from synthetic construct)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Histone h2a (11 kDa, Protein from Homo sapiens)
- Histone-lysine n-methyltransferase, h3 lysine-79 specific (37 kDa, Protein from Homo sapiens)
- Histone h4 (10 kDa, Protein from Homo sapiens)
- S-adenosylmethionine (398 Da, Ligand)
Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex
Resolution: 2.72 Å
EM Method: Single-particle
Fitted PDBs: 7xct
Q-score: 0.514
Ai H, Sun M, Liu A, Sun Z, Liu T, Cao L, Liang L, Qu Q, Li Z, Deng Z, Tong Z, Chu G, Tian X, Deng H, Zhao S, Li JB, Lou Z, Liu L
Nat Chem Biol (2022) 18 pp. 972-980 [ Pubmed: 35739357 DOI: doi:10.1038/s41589-022-01067-7 ]
- Histone domain-containing protein (11 kDa, Protein from Homo sapiens)
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Complex of dot1l(1-416) and h2bk34ub 2:1 nucleosome (Complex from Homo sapiens)
- Dna (145-mer) (44 kDa, DNA from synthetic construct)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Histone h2a (11 kDa, Protein from Homo sapiens)
- Dna (145-mer) (44 kDa, DNA from synthetic construct)
- Histone h4 (10 kDa, Protein from Homo sapiens)
- Histone-lysine n-methyltransferase, h3 lysine-79 specific (37 kDa, Protein from Homo sapiens)
- S-adenosylmethionine (398 Da, Ligand)
cryo-EM map of Dot1L and H2BK34ub-H3K79Nle nucleosome complex containing addition map (1:1)
SARS-CoV-2 spike in complex with LCB1 (2RBDs open)
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7jzl
Q-score: 0.493
Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park YJ, Strauch EM, Stewart L, Diamond MS, Veesler D, Baker D
Science (2020) 370 pp. 426-431 [ Pubmed: 32907861 DOI: doi:10.1126/science.abd9909 ]
- Lcb1 (Complex from synthetic construct)
- Lcb1 (6 kDa, Protein from synthetic construct)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- De novo design of picomolar sars-cov-2 miniprotein inhibitors (Complex)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors in reduced conditions
Resolution: 2.95 Å
EM Method: Single-particle
Fitted PDBs: 7btf
Q-score: 0.569
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z
Science (2020) 368 pp. 779-782 [ Pubmed: 32277040 DOI: doi:10.1126/science.abb7498 ]
- Rna-directed rna polymerase (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Structure of sars-cov-2 rna-dependent rna polymerase in complex with cofactors in reduced condition (155 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)