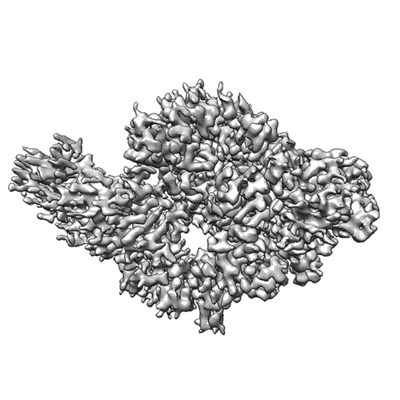
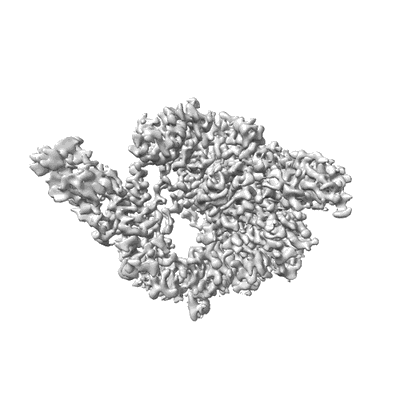
Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex
Resolution: 2.57 Å
EM Method: Single-particle
Fitted PDBs: 7xcr
Q-score: 0.571
Ai H, Sun M, Liu A, Sun Z, Liu T, Cao L, Liang L, Qu Q, Li Z, Deng Z, Tong Z, Chu G, Tian X, Deng H, Zhao S, Li JB, Lou Z, Liu L
Nat Chem Biol (2022) 18 pp. 972-980 [ Pubmed: 35739357 DOI: doi:10.1038/s41589-022-01067-7 ]
- Histone h4 (10 kDa, Protein from Homo sapiens)
- Complex of dot1l(1-416) and h2bk34ub nucleosome (Complex from Homo sapiens)
- Histone-lysine n-methyltransferase, h3 lysine-79 specific (37 kDa, Protein from Homo sapiens)
- Dna (146-mer) (44 kDa, DNA from synthetic construct)
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- S-adenosylmethionine (398 Da, Ligand)
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Dna (146-mer) (45 kDa, DNA from synthetic construct)
- Histone domain-containing protein (11 kDa, Protein from synthetic construct)
- Histone h2a (11 kDa, Protein from Homo sapiens)
Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex
Resolution: 2.72 Å
EM Method: Single-particle
Fitted PDBs: 7xct
Q-score: 0.514
Ai H, Sun M, Liu A, Sun Z, Liu T, Cao L, Liang L, Qu Q, Li Z, Deng Z, Tong Z, Chu G, Tian X, Deng H, Zhao S, Li JB, Lou Z, Liu L
Nat Chem Biol (2022) 18 pp. 972-980 [ Pubmed: 35739357 DOI: doi:10.1038/s41589-022-01067-7 ]
- Histone h2b type 1-k (10 kDa, Protein from Homo sapiens)
- Dna (145-mer) (44 kDa, DNA from synthetic construct)
- Histone-lysine n-methyltransferase, h3 lysine-79 specific (37 kDa, Protein from Homo sapiens)
- Complex of dot1l(1-416) and h2bk34ub 2:1 nucleosome (Complex from Homo sapiens)
- S-adenosylmethionine (398 Da, Ligand)
- Ubiquitin (8 kDa, Protein from Homo sapiens)
- Histone domain-containing protein (11 kDa, Protein from Homo sapiens)
- Histone h4 (10 kDa, Protein from Homo sapiens)
- Dna (145-mer) (44 kDa, DNA from synthetic construct)
- Histone h2a (11 kDa, Protein from Homo sapiens)
cryo-EM map of Dot1L and H2BK34ub-H3K79Nle nucleosome complex containing addition map (1:1)
SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors in reduced conditions
Resolution: 2.95 Å
EM Method: Single-particle
Fitted PDBs: 7btf
Q-score: 0.569
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z
Science (2020) 368 pp. 779-782 [ Pubmed: 32277040 DOI: doi:10.1126/science.abb7498 ]
- Rna-directed rna polymerase (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Structure of sars-cov-2 rna-dependent rna polymerase in complex with cofactors in reduced condition (155 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6m71
Q-score: 0.519
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z
Science (2020) 368 pp. 779-782 [ Pubmed: 32277040 DOI: doi:10.1126/science.abb7498 ]
- Rna-directed rna polymerase (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 rna-dependent rna polymerase in complex with cofactors (155 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Anthrax Toxin Receptor 1-bound full particles of Seneca Valley Virus in acidic conditions
Resolution: 2.84 Å
EM Method: Single-particle
Fitted PDBs: 6adm
Q-score: -0.027
Cao L, Zhang R, Liu T, Sun Z, Hu M, Sun Y, Cheng L, Guo Y, Fu S, Hu J, Li X, Yu C, Wang H, Chen H, Li X, Fry EE, Stuart DI, Qian P, Lou Z, Rao Z
PNAS (2018) 115 pp. 13087-13092 [ Pubmed: 30514821 DOI: doi:10.1073/pnas.1814309115 ]
- Anthrax toxin receptor 1 (21 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Seneca valley virus (Virus from Seneca valley virus)
- Vp3 (26 kDa, Protein from Seneca valley virus)
- Vp4 (5 kDa, Protein from Seneca valley virus)
- Vp1 (28 kDa, Protein from Seneca valley virus)
- Vp2 (29 kDa, Protein from Seneca valley virus)
Structure of Seneca Valley Virus in acidic conditions
Resolution: 2.84 Å
EM Method: Single-particle
Fitted PDBs: 6ads
Q-score: -0.019
Cao L, Zhang R, Liu T, Sun Z, Hu M, Sun Y, Cheng L, Guo Y, Fu S, Hu J, Li X, Yu C, Wang H, Chen H, Li X, Fry EE, Stuart DI, Qian P, Lou Z, Rao Z
PNAS (2018) 115 pp. 13087-13092 [ Pubmed: 30514821 DOI: doi:10.1073/pnas.1814309115 ]
- Vp3 (25 kDa, Protein from Seneca valley virus)
- Vp4 (7 kDa, Protein from Seneca valley virus)
- Seneca valley virus (Virus from Seneca valley virus)
- Vp2 (29 kDa, Protein from Seneca valley virus)
- Vp1 (28 kDa, Protein from Seneca valley virus)