Motion corrected micrographs - purified SINV/EEEV particles and recombinant anti-EEEV Fab (EEEV-143)
Williamson LE, Gilliland T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe JE
Cell (2020) 183 pp. 1884-1.9e+26 [ Pubmed: 33301709 DOI: 10.1016/j.cell.2020.11.011 ]
Image sets:
Motion corrected micrographs - purified SINV/EEEV particles and recombinant anti-EEEV Fab (EEEV-33)
Williamson LE, Gilliland T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: 10.1016/j.cell.2020.11.011 ]
Image sets:
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-143 Antibody
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-33 Antibody
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
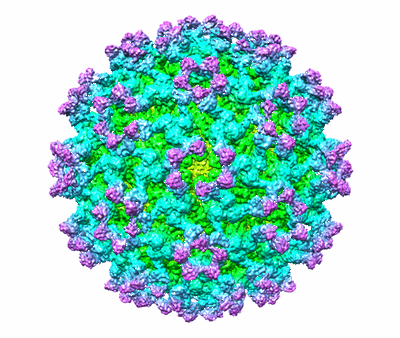
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 6xo4
Q-score: 0.343
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
- Eastern equine encephalitis virus (Virus from Eastern equine encephalitis virus)
- Togavirin (29 kDa, Protein from Eastern equine encephalitis virus)
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP with Fab EEEV-143.
Resolution: 8.5 Å
EM Method: Single-particle
Fitted PDBs: 6xob
Q-score: 0.182
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
- Eastern equine encephalitis virus (Virus from Eastern equine encephalitis virus)
- Togavirin (29 kDa, Protein from Eastern equine encephalitis virus)
- Eeev-143 fab light chain (23 kDa, Protein from Homo sapiens)
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
- Eeev-143 fab heavy chain (23 kDa, Protein from Homo sapiens)
Negative stain EM map of SARS CoV-2 spike protein (trimer)
Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo YM, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe Jr JE
Nature (2020) 584 pp. 443-449 [ DOI: doi:10.1038/s41586-020-2548-6 Pubmed: 32668443 ]
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2165
Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo YM, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe Jr JE
Nature (2020) 584 pp. 443-449 [ DOI: doi:10.1038/s41586-020-2548-6 Pubmed: 32668443 ]
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2196
Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo YM, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe Jr JE
Nature (2020) 584 pp. 443-449 [ DOI: doi:10.1038/s41586-020-2548-6 Pubmed: 32668443 ]
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2130
Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo YM, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe Jr JE
Nature (2020) 584 pp. 443-449 [ DOI: doi:10.1038/s41586-020-2548-6 Pubmed: 32668443 ]