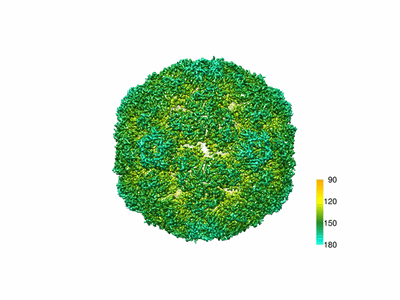
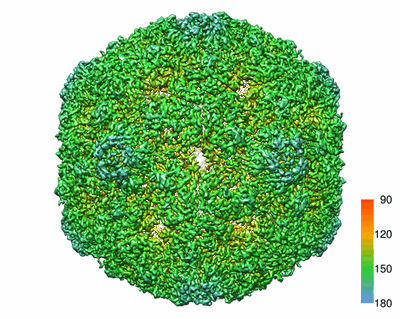
Resolution: 4.176 Å
EM Method: Single-particle
Fitted PDBs: 7ymd
Q-score: 0.319
Li Q, Zhang J, Haluska C, Zhang X, Wang L, Liu G, Wang Z, Jin D, Cheng T, Wang H, Tian Y, Wang X, Sun L, Zhao X, Chen Z, Wang L
Nat Struct Mol Biol (2024) [ Pubmed: 38890552 DOI: doi:10.1038/s41594-024-01319-1 ]
- Non-structural maintenance of chromosome element 4 (46 kDa, Protein from Saccharomyces cerevisiae S288C)
- Non-structural maintenance of chromosome element 3 (34 kDa, Protein from Saccharomyces cerevisiae S288C)
- Cryo-em structure of nse1/3/4 (Complex from Saccharomyces cerevisiae S288c)
- Non-structural maintenance of chromosomes element 1 (38 kDa, Protein from Saccharomyces cerevisiae S288C)
The atomic structure of varicella-zoster virus A-capsid
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 6lgl
Q-score: 0.114
Wang W, Zheng Q, Pan D, Yu H, Fu W, Liu J, He M, Zhu R, Cai Y, Huang Y, Zha Z, Chen Z, Ye X, Han J, Que Y, Wu T, Zhang J, Li S, Zhu H, Zhou ZH, Cheng T, Xia N
Nat Microbiol (2020) 5 pp. 1542-1552 [ DOI: doi:10.1038/s41564-020-0785-y Pubmed: 32895526 ]
- Triplex capsid protein 2 (34 kDa, Protein from Human herpesvirus 3)
- Major capsid protein (155 kDa, Protein from Human herpesvirus 3)
- Triplex capsid protein 1 (54 kDa, Protein from Human herpesvirus 3)
- Human alphaherpesvirus 3 (Virus from Human alphaherpesvirus 3)
- Small capsomere-interacting protein (24 kDa, Protein from Human herpesvirus 3)
The structure of CVA10 virus procapsid particle
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6acw
Q-score: 0.476
Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, Li Z, Chen Z, Yu H, Que Y, Liu C, Kong Z, Zhang J, Baker TS, Yan X, Hong Zhou Z, Cheng T, Xia N
Sci Adv (2018) 4 pp. eaat7459-eaat7459 [ DOI: doi:10.1126/sciadv.aat7459 Pubmed: 30255146 ]
- Vp0 (35 kDa, Protein from Coxsackievirus A10)
- Coxsackievirus a10 (Virus from Coxsackievirus A10)
- Vp3 (26 kDa, Protein from Coxsackievirus A10)
- Vp1 (33 kDa, Protein from Coxsackievirus A10)
The structure of CVA10 virus A-particle from its complex with Fab 2G8
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 6acz
Q-score: 0.466
Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, Li Z, Chen Z, Yu H, Que Y, Liu C, Kong Z, Zhang J, Baker TS, Yan X, Hong Zhou Z, Cheng T, Xia N
Sci Adv (2018) 4 pp. eaat7459-eaat7459 [ DOI: doi:10.1126/sciadv.aat7459 Pubmed: 30255146 ]
- Vp2 (27 kDa, Protein from Coxsackievirus A10)
- Coxsackievirus a10 (Virus from Coxsackievirus A10)
- Vp3 (26 kDa, Protein from Coxsackievirus A10)
- Vp1 (33 kDa, Protein from Coxsackievirus A10)
The structure of CVA10 procapsid from its complex with Fab 2G8
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 6ad1
Q-score: 0.481
Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, Li Z, Chen Z, Yu H, Que Y, Liu C, Kong Z, Zhang J, Baker TS, Yan X, Hong Zhou Z, Cheng T, Xia N
Sci Adv (2018) 4 pp. eaat7459-eaat7459 [ DOI: doi:10.1126/sciadv.aat7459 Pubmed: 30255146 ]
- Vp0 (35 kDa, Protein from Coxsackievirus A10)
- Coxsackievirus a10 (Virus from Coxsackievirus A10)
- Vp3 (26 kDa, Protein from Coxsackievirus A10)
- Vp1 (33 kDa, Protein from Coxsackievirus A10)
The structure of ICAM-5 triggered Enterovirus D68 virus A-particle
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6aj2
Q-score: 0.412
Zheng Q, Zhu R, Xu L, He M, Yan X, Liu D, Yin Z, Wu Y, Li Y, Yang L, Hou W, Li S, Li Z, Chen Z, Li Z, Yu H, Gu Y, Zhang J, Baker TS, Zhou ZH, Graham BS, Cheng T, Li S, Xia N
Nat Microbiol (2019) 4 pp. 124-133 [ Pubmed: 30397341 DOI: doi:10.1038/s41564-018-0275-7 ]
- Capsid protein vp3 (27 kDa, Protein from Enterovirus D68)
- Capsid protein vp1 (32 kDa, Protein from Enterovirus D68)
- Capsid protein vp2 (27 kDa, Protein from Enterovirus D68)
- Enterovirus d (Virus from Enterovirus D)