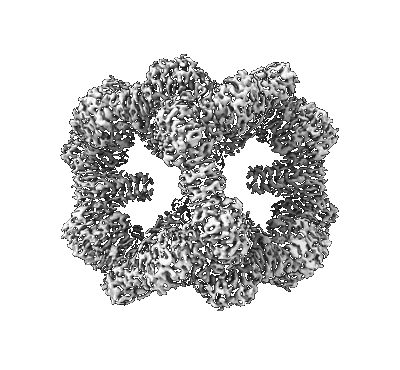
Accurate computational design of genetically encoded 3D protein crystals
Resolution: 3.34 Å
EM Method: Single-particle
Fitted PDBs: 8cwy
Q-score: 0.452
Li Z, Wang S, Nattermann U, Bera AK, Borst AJ, Yaman MY, Bick MJ, Yang EC, Sheffler W, Lee B, Seifert S, Hura GL, Nguyen H, Kang A, Dalal R, Lubner JM, Hsia Y, Haddox H, Courbet A, Dowling Q, Miranda M, Favor A, Etemadi A, Edman NI, Yang W, Weidle C, Sankaran B, Negahdari B, Ross MB, Ginger DS, Baker D
Nat Mater (2023) 22 pp. 1556-1563 [ Pubmed: 37845322 DOI: doi:10.1038/s41563-023-01683-1 ]
- T32-15-1 (49 kDa, Protein from synthetic construct)
- T32-15 (Complex from Escherichia coli)
- T32-15-2 (9 kDa, Protein from synthetic construct)
CryoEM Structure of Computationally Designed Nanocage O32-ZL4
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8szz
Q-score: 0.591
Li Z, Wang S, Nattermann U, Bera AK, Borst AJ, Yaman MY, Bick MJ, Yang EC, Sheffler W, Lee B, Seifert S, Hura GL, Nguyen H, Kang A, Dalal R, Lubner JM, Hsia Y, Haddox H, Courbet A, Dowling Q, Miranda M, Favor A, Etemadi A, Edman NI, Yang W, Weidle C, Sankaran B, Negahdari B, Ross MB, Ginger DS, Baker D
Nat Mater (2023) 22 pp. 1556-1563 [ Pubmed: 37845322 DOI: doi:10.1038/s41563-023-01683-1 ]
- O32-zl4 component a (8 kDa, Protein from synthetic construct)
- O32-zl4 (764 kDa, Complex from Thermotoga maritima)
- Sodium ion (22 Da, Ligand)
- O32-zl4 component b (23 kDa, Protein from Thermotoga maritima)
CryoEM map of a de novo designed octahedral nanocage with programmable volume; design cage_O4_34
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Weidle C, Han HL, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
bioRxiv (2023) [ Pubmed: 37333359 DOI: doi:10.1101/2023.06.09.544258 ]
- Cage_o4_34 (Complex from synthetic construct)
- Cage_o4_34 (Protein from synthetic construct)
Cryo-EM map of synthetic cage_O3_10 reconstructed without symmetry (C1)
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
Cryo-EM map of synthetic cage_O3_10 reconstructed with O symmetry
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Weidle C, Han HL, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
bioRxiv (2023) [ DOI: doi:10.1101/2023.06.09.544258 Pubmed: 37333359 ]
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
Cryo-EM map of synthetic cage_T3_5 reconstructed with T symmetry
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
Cryo-EM map of synthetic cage_T3_5 reconstructed without symmetry (C1)
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
Cryo-EM map of synthetic cage_T3_5+2 reconstructed without symmetry (C1)
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]