McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
Proc Natl Acad Sci U S A (2023) 120 [ Pubmed: 38011573 DOI: 10.1073/pnas.2312905120 ]
Image sets:
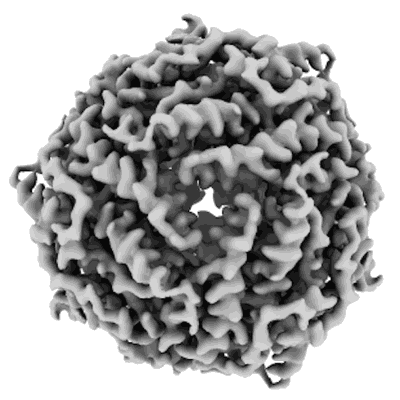
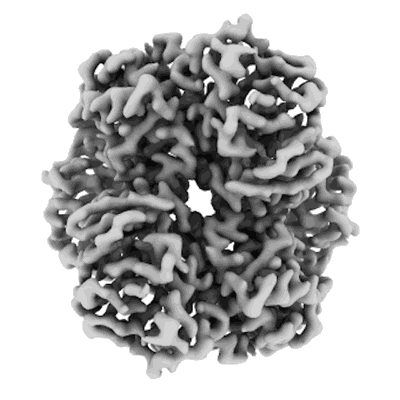
Structure of DPS determined by cryoEM at 100 keV
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8pv9
Q-score: 0.615
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
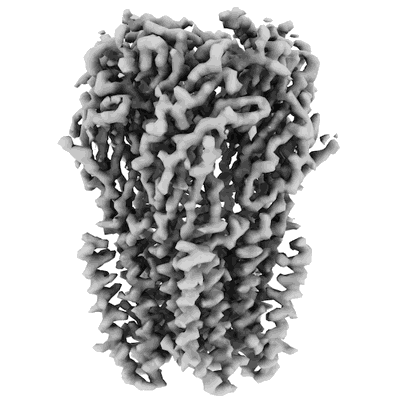
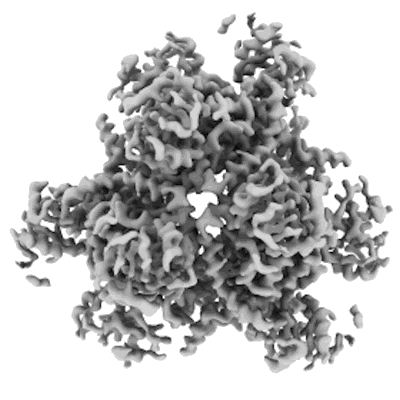
Structure of bacterial ribosome determined by cryoEM at 100 keV
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 8pva
Q-score: 0.287
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
- Small ribosomal subunit protein us8 (14 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l17 (14 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l19 (13 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l14 (13 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l15 (15 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l23 (11 kDa, Protein from Escherichia coli)
- E. coli 70s ribosome (Complex from Escherichia coli)
- Large ribosomal subunit protein ul4 (22 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l35 (7 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us15 (10 kDa, Protein from Escherichia coli)
- 23s rrna (941 kDa, RNA from Escherichia coli)
- Large ribosomal subunit protein ul2 (29 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l33 (6 kDa, Protein from Escherichia coli)
- Large ribosomal subunit protein ul6 (18 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l20 (13 kDa, Protein from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- 50s ribosomal protein l29 (7 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us9 (14 kDa, Protein from Escherichia coli)
- 16s rrna (499 kDa, RNA from Escherichia coli)
- Mrna (9 kDa, RNA from Escherichia coli)
- Small ribosomal subunit protein us5 (17 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l18 (12 kDa, Protein from Escherichia coli)
- E-site trna (589 Da, RNA from Escherichia coli)
- Small ribosomal subunit protein us11 (13 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l21 (11 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein bs16 (9 kDa, Protein from Escherichia coli)
- Large ribosomal subunit protein bl9 (15 kDa, Protein from Escherichia coli)
- Large ribosomal subunit protein ul3 (22 kDa, Protein from Escherichia coli)
- Large ribosomal subunit protein ul16 (15 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l36 (4 kDa, Protein from Escherichia coli)
- P-site trna-fmet (24 kDa, RNA from Escherichia coli)
- 50s ribosomal protein l27 (9 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us19 (10 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us2 (26 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein bs20 (9 kDa, Protein from Escherichia coli)
- Large ribosomal subunit protein ul13 (16 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l32 (6 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein bs21 (8 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us10 (11 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein bs6, fully modified isoform (15 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us4 (23 kDa, Protein from Escherichia coli)
- Large ribosomal subunit protein bl31a (7 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l22 (12 kDa, Protein from Escherichia coli)
- 5s rrna (38 kDa, RNA from Escherichia coli)
- 50s ribosomal protein l24 (11 kDa, Protein from Escherichia coli)
- A-site trna-val (24 kDa, RNA from Escherichia coli)
- Small ribosomal subunit protein us13 (13 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l30 (6 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us17 (9 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein bs18 (9 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l28 (9 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us3 (26 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l25 (10 kDa, Protein from Escherichia coli)
- 50s ribosomal protein l34 (5 kDa, Protein from Escherichia coli)
- Large ribosomal subunit protein ul5 (20 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us12 (13 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us14 (11 kDa, Protein from Escherichia coli)
- Small ribosomal subunit protein us7 (20 kDa, Protein from Escherichia coli)
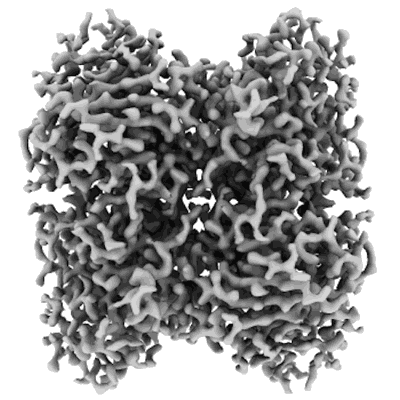
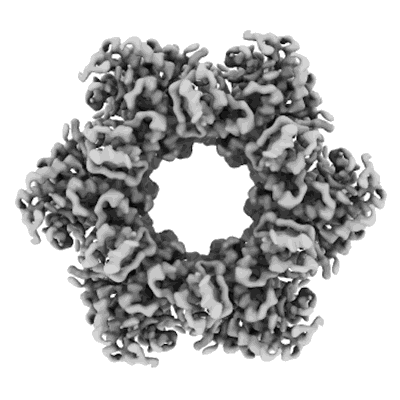
Structure of GABAAR determined by cryoEM at 100 keV
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8pvb
Q-score: 0.486
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ Pubmed: 38011573 DOI: doi:10.1073/pnas.2312905120 ]
- Decane (142 Da, Ligand)
- Megabody mb25 (56 kDa, Protein from Lama glama)
- Histamine (111 Da, Ligand)
- Hexadecane (226 Da, Ligand)
- Human gaba(a)r-beta3 homopentamer in complex with megabody-25 in lipid nanodisc (Complex from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Gamma-aminobutyric acid receptor subunit beta-3 (45 kDa, Protein from Homo sapiens)
- Etomidate (244 Da, Ligand)
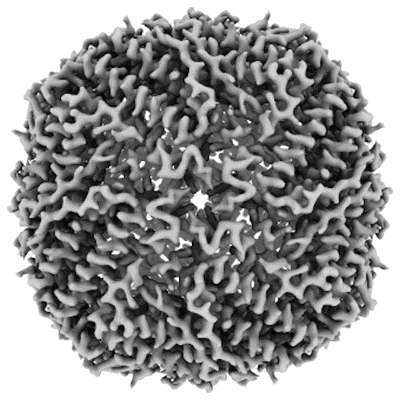
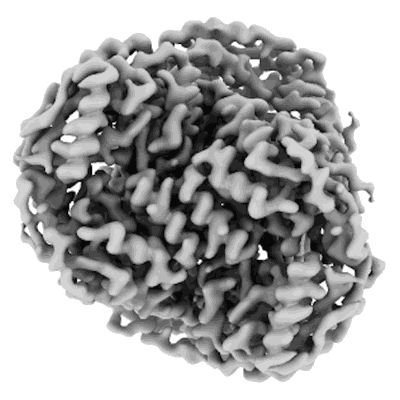
Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8pvc
Q-score: 0.675
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
- Ferritin heavy chain (21 kDa, Protein from Mus musculus)
- Zinc ion (65 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Mouse heavy-chain apoferritin (Complex from Mus musculus)
Structure of catalase determined by cryoEM at 100 keV
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8pvd
Q-score: 0.523
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Nadph dihydro-nicotinamide-adenine-dinucleotide phosphate (745 Da, Ligand)
- Catalase (59 kDa, Protein from Homo sapiens)
- Catalase from human erythrocytes (Complex from Homo sapiens)
Structure of AHIR determined by cryoEM at 100 keV
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8pve
Q-score: 0.526
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
Structure of GAPDH determined by cryoEM at 100 keV
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8pvf
Q-score: 0.572
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8pvg
Q-score: 0.463
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
Structure of human apo ALDH1A1 determined by cryoEM at 100 keV
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8pvh
Q-score: 0.55
McMullan G, Naydenova K, Mihaylov D, Yamashita K, Peet MJ, Wilson H, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy MA, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
PNAS (2023) 120 pp. e2312905120-e2312905120 [ DOI: doi:10.1073/pnas.2312905120 Pubmed: 38011573 ]
- Chloride ion (35 Da, Ligand)
- Aldehyde dehydrogenase 1a1 (57 kDa, Protein from Homo sapiens)
- Human apo aldh1a1 (Complex from Homo sapiens)