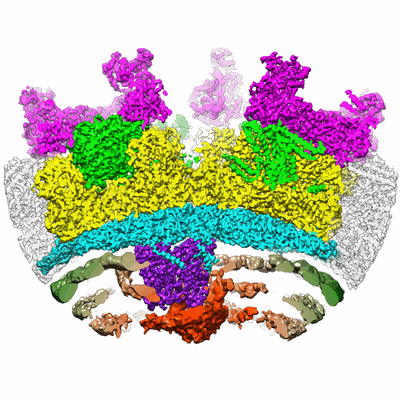
Cryo-EM structure of murine Dispatched 'R' conformation
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7rph
Q-score: 0.57
Wang Q, Asarnow DE, Ding K, Mann RK, Hatakeyama J, Zhang Y, Ma Y, Cheng Y, Beachy PA
Nature (2021) 599 pp. 320-324 [ Pubmed: 34707294 DOI: doi:10.1038/s41586-021-03996-0 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Lauryl maltose neopentyl glycol (1 kDa, Ligand)
- Dispatched protein 'r' conformation (151 kDa, Complex from Mus musculus)
- Sodium ion (22 Da, Ligand)
- Protein dispatched homolog 1 (152 kDa, Protein from Mus musculus)
- Cholesterol hemisuccinate (486 Da, Ligand)
- Water (18 Da, Ligand)
Cryo-EM structure of murine Dispatched 'T' conformation
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7rpi
Q-score: 0.554
Wang Q, Asarnow DE, Ding K, Mann RK, Hatakeyama J, Zhang Y, Ma Y, Cheng Y, Beachy PA
Nature (2021) 599 pp. 320-324 [ Pubmed: 34707294 DOI: doi:10.1038/s41586-021-03996-0 ]
- Dispatched protein 't' conformation (151 kDa, Complex from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Lauryl maltose neopentyl glycol (1 kDa, Ligand)
- Sodium ion (22 Da, Ligand)
- Protein dispatched homolog 1 (152 kDa, Protein from Mus musculus)
- Cholesterol hemisuccinate (486 Da, Ligand)
- Water (18 Da, Ligand)
Cryo-EM structure of murine Dispatched NNN mutant
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7rpj
Q-score: 0.427
Wang Q, Asarnow DE, Ding K, Mann RK, Hatakeyama J, Zhang Y, Ma Y, Cheng Y, Beachy PA
Nature (2021) 599 pp. 320-324 [ Pubmed: 34707294 DOI: doi:10.1038/s41586-021-03996-0 ]
- Cholesterol hemisuccinate (486 Da, Ligand)
- Dispatched protein nnn mutant (151 kDa, Complex from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Protein dispatched homolog 1 (152 kDa, Protein from Mus musculus)
Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7rpk
Q-score: 0.547
Wang Q, Asarnow DE, Ding K, Mann RK, Hatakeyama J, Zhang Y, Ma Y, Cheng Y, Beachy PA
Nature (2021) 599 pp. 320-324 [ Pubmed: 34707294 DOI: doi:10.1038/s41586-021-03996-0 ]
- Zinc ion (65 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Lauryl maltose neopentyl glycol (1 kDa, Ligand)
- Sulfate ion (96 Da, Ligand)
- Dispatched protein complexed with sonic hedgehog 26-189 (151 kDa, Complex from Mus musculus)
- Sodium ion (22 Da, Ligand)
- Sonic hedgehog protein (18 kDa, Protein from Mus musculus)
- Protein dispatched homolog 1 (152 kDa, Protein from Mus musculus)
- Cholesterol hemisuccinate (486 Da, Ligand)
- (2s)-3-{[(s)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate (411 Da, Ligand)
- Water (18 Da, Ligand)
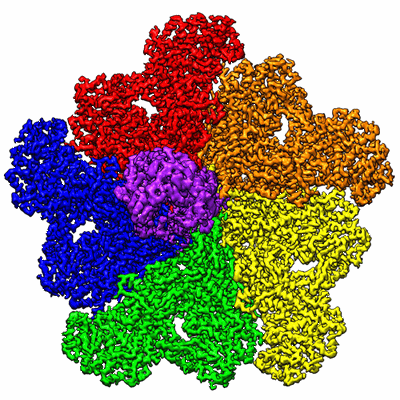
In situ structure of BTV RNA-dependent RNA polymerase in BTV virion
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6pns
Q-score: 0.484
He Y, Shivakoti S, Ding K, Cui Y, Roy P, Zhou ZH
PNAS (2019) 116 pp. 16535-16540 [ DOI: doi:10.1073/pnas.1905849116 Pubmed: 31350350 ]
- Inner core structural protein vp3 (103 kDa, Protein from Bluetongue virus 1)
- Bluetongue virus 1 (Virus from Bluetongue virus 1)
- Rna-directed rna polymerase (149 kDa, Protein from Bluetongue virus 1)
In situ structure of BTV RNA-dependent RNA polymerase in BTV core
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 6po2
Q-score: 0.497
He Y, Shivakoti S, Ding K, Cui Y, Roy P, Zhou ZH
PNAS (2019) 116 pp. 16535-16540 [ DOI: doi:10.1073/pnas.1905849116 Pubmed: 31350350 ]
- Inner core structural protein vp3 (103 kDa, Protein from Bluetongue virus 1)
- Bluetongue virus 1 (Virus from Bluetongue virus 1)
- Rna-directed rna polymerase (149 kDa, Protein from Bluetongue virus 1)
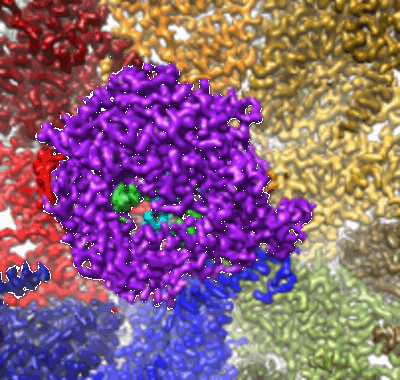
In situ structure of Rotavirus RNA-dependent RNA polymerase at duplex-open state
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6ogy
Q-score: 0.537
Ding K, Celma CC, Zhang X, Chang T, Shen W, Atanasov I, Roy P, Zhou ZH
Nat Commun (2019) 10 pp. 2216-2216 [ Pubmed: 31101900 DOI: doi:10.1038/s41467-019-10236-7 ]
- Rna-dependent rna polymerase of rotavirus a (125 kDa, Protein from Rotavirus A)
- Rotavirus a (Virus from Rotavirus A)
- Dna/rna (5'-d(*(gtg))-r(p*gp*c)-3') (1 kDa, Macromolecule from Rotavirus A)
- Rna (5'-r(p*ap*gp*cp*c)-3') (1 kDa, RNA from Rotavirus A)
- Inner capsid protein vp2 (103 kDa, Protein from Rotavirus A)
In situ structure of Rotavirus RNA-dependent RNA polymerase at transcript-elongated state
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 6ogz
Q-score: 0.517
Ding K, Celma CC, Zhang X, Chang T, Shen W, Atanasov I, Roy P, Zhou ZH
Nat Commun (2019) 10 pp. 2216-2216 [ Pubmed: 31101900 DOI: doi:10.1038/s41467-019-10236-7 ]
- Uridine 5'-triphosphate (484 Da, Ligand)
- Rna (5'-r(p*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*a)-3') (5 kDa, RNA from Rotavirus A)
- Rna-dependent rna polymerase of rotavirus a (125 kDa, Protein from Rotavirus A)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Rotavirus a (Virus from Rotavirus A)
- Rna (5'-r(p*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*a)-3') (5 kDa, RNA from Rotavirus A)
- Inner capsid protein vp2 (103 kDa, Protein from Rotavirus A)
Recombinant major vault protein [Rattus norvegicus] structure in solution: conformation 2
Resolution: 4.9 Å
EM Method: Single-particle
Fitted PDBs: 6bp7
Q-score: 0.311
Recombinant major vault protein [Rattus norvegicus] structure in solution: conformation 1
Resolution: 4.9 Å
EM Method: Single-particle
Fitted PDBs: 6bp8
Q-score: 0.289