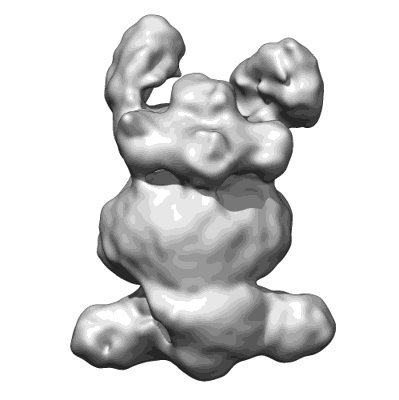
Mycobacterium tuberculosis CoaX Homohexamer
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 9b78
Q-score: 0.579
Mycobacterium tuberculosis CoaX Homotetramer
Resolution: 2.71 Å
EM Method: Single-particle
Fitted PDBs: 9b79
Q-score: 0.522
Complex of M. smegmatis Dop with M. tuberculosis CoaX and Pup91 (Composite Map)
Resolution: 2.04 Å
EM Method: Single-particle
Fitted PDBs: 9cku
Q-score: 0.733
Kahne SC, Yoo JH, Chen J, Nakedi K, Iyer LM, Putzel G, Samhadaneh NM, Pironti A, Aravind L, Ekiert DC, Bhabha G, Rhee KY, Darwin KH
PNAS (2024) 121 pp. e2407239121-e2407239121 [ Pubmed: 39585979 DOI: doi:10.1073/pnas.2407239121 ]
- Water (18 Da, Ligand)
- Complex of m. smegmatis dop with m. tuberculosis coax and pup91 (Complex from Mycobacterium tuberculosis)
- Type iii pantothenate kinase (29 kDa, Protein from Mycobacterium tuberculosis)
- Pup (7 kDa, Protein from Mycobacterium tuberculosis)
- Pup deamidase/depupylase (54 kDa, Protein from Mycolicibacterium smegmatis)
MacRae MR, Puvanendran D, Haase MAB, Coudray N, Kolich L, Lam C, Baek M, Bhabha G, Ekiert DC
J Biol Chem (2023) 299 pp. 104744-104744 [ DOI: doi:10.1016/j.jbc.2023.104744 Pubmed: 37100290 ]
- Mlae (Protein from Escherichia coli K-12)
- Mlad (Protein from Escherichia coli K-12)
- E. coli mlac bound to mlafedb (Complex from Escherichia coli K-12)
- Foldon-mlac (Protein from Escherichia coli K-12)
- Mlab (Protein from Escherichia coli K-12)
- Mlaf (Protein from Escherichia coli K-12)
MacRae MR, Puvanendran D, Haase MAB, Coudray N, Kolich L, Lam C, Baek M, Bhabha G, Ekiert DC
J Biol Chem (2023) 299 pp. 104744-104744 [ DOI: doi:10.1016/j.jbc.2023.104744 Pubmed: 37100290 ]
- E. coli mlafedb bound to mlac (Complex from Escherichia coli K-12)
- Mlae (Protein from Escherichia coli K-12)
- Mlad (Protein from Escherichia coli K-12)
- Mlac-foldon (Protein from Escherichia coli K-12)
- Mlab (Protein from Escherichia coli K-12)
- Mlaf (Protein from Escherichia coli K-12)
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7mi3
Q-score: 0.441
Santarossa CC, Mickolajczyk KJ, Steinman JB, Urnavicius L, Chen N, Hirata Y, Fukase Y, Coudray N, Ekiert DC, Bhabha G, Kapoor TM
Cell Chem Biol (2021) 28 pp. 1460-1473.e15 [ Pubmed: 34015309 DOI: doi:10.1016/j.chembiol.2021.04.024 ]
- Dynein motor domain in the presence of a pyrazolo-pyrimidinone-based compound (300 kDa, Complex from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Fusion protein of dynein and endolysin (304 kDa, Protein from Saccharomyces cerevisiae)
- (8s)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile (481 Da, Ligand)
Yeast dynein motor domain in the presence of a pyrazolo-pyrimidinone-based compound, Map 1
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7mi6
Q-score: 0.339
Santarossa CC, Mickolajczyk KJ, Steinman JB, Urnavicius L, Chen N, Hirata Y, Fukase Y, Coudray N, Ekiert DC, Bhabha G, Kapoor TM
Cell Chem Biol (2021) 28 pp. 1460-1473.e15 [ Pubmed: 34015309 DOI: doi:10.1016/j.chembiol.2021.04.024 ]
- Dynein motor domain in the presence of a pyrazolo-pyrimidinone-based compound (300 kDa, Complex from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Fusion protein of dynein and endolysin (304 kDa, Protein from Saccharomyces cerevisiae)
- (8s)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile (481 Da, Ligand)
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 7mi8
Q-score: 0.427
Santarossa CC, Mickolajczyk KJ, Steinman JB, Urnavicius L, Chen N, Hirata Y, Fukase Y, Coudray N, Ekiert DC, Bhabha G, Kapoor TM
Cell Chem Biol (2021) 28 pp. 1460-1473.e15 [ Pubmed: 34015309 DOI: doi:10.1016/j.chembiol.2021.04.024 ]