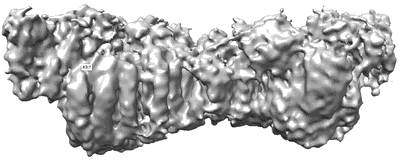
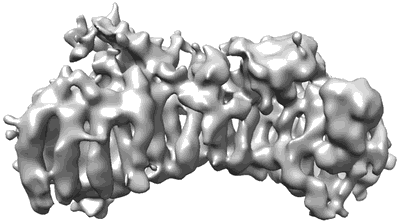
Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 7ao8
Q-score: 0.105
Millard CJ, Fairall L, Ragan TJ, Savva CG, Schwabe JWR
Nucleic Acids Res (2020) 48 pp. 12972-12982 [ Pubmed: 33264408 DOI: doi:10.1093/nar/gkaa1121 ]
- Zinc ion (65 Da, Ligand)
- Nurd deacetylase complex containing two copies of mta1 and hdac1 and a single copy of mbd2 (220 kDa, Complex from Homo sapiens)
- Methyl-cpg-binding domain protein 2 (43 kDa, Protein from Homo sapiens)
- Potassium ion (39 Da, Ligand)
- Metastasis-associated protein mta1 (80 kDa, Protein from Homo sapiens)
- Inositol hexakisphosphate (660 Da, Ligand)
- Histone deacetylase 1 (55 kDa, Protein from Homo sapiens)
Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex
Resolution: 6.1 Å
EM Method: Single-particle
Fitted PDBs: 7ao9
Q-score: 0.13
Millard CJ, Fairall L, Ragan TJ, Savva CG, Schwabe JWR
Nucleic Acids Res (2020) 48 pp. 12972-12982 [ Pubmed: 33264408 DOI: doi:10.1093/nar/gkaa1121 ]
- Zinc ion (65 Da, Ligand)
- Methyl-cpg-binding domain protein 2 (43 kDa, Protein from Homo sapiens)
- Potassium ion (39 Da, Ligand)
- Metastasis-associated protein mta1 (80 kDa, Protein from Homo sapiens)
- Inositol hexakisphosphate (660 Da, Ligand)
- Histone deacetylase 1 (55 kDa, Protein from Homo sapiens)
- Core nurd deacetylase complex containing two copies of mta1 and hdac1 and a single copy of mbd2 (170 kDa, Complex from Homo sapiens)
Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex
Resolution: 19.4 Å
EM Method: Single-particle
Fitted PDBs: 7aoa
Q-score: 0.06
Millard CJ, Fairall L, Ragan TJ, Savva CG, Schwabe JWR
Nucleic Acids Res (2020) 48 pp. 12972-12982 [ Pubmed: 33264408 DOI: doi:10.1093/nar/gkaa1121 ]
- Zinc ion (65 Da, Ligand)
- Methyl-cpg-binding domain protein 2 (43 kDa, Protein from Homo sapiens)
- Histone-binding protein rbbp4 (47 kDa, Protein from Homo sapiens)
- Potassium ion (39 Da, Ligand)
- Inositol hexakisphosphate (660 Da, Ligand)
- Metastasis-associated protein mta1 (80 kDa, Protein from Homo sapiens)
- Extended nurd deacetylase complex containing two copies of mta1, hdac1 and rbbp4 and a single copy of mbd2 (340 kDa, Complex from Homo sapiens)
- Histone deacetylase 1 (55 kDa, Protein from Homo sapiens)
The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6z2j
Q-score: 0.314
Turnbull RE, Fairall L, Saleh A, Kelsall E, Morris KL, Ragan TJ, Savva CG, Chandru A, Millard CJ, Makarova OV, Smith CJ, Roseman AM, Fry AM, Cowley SM, Schwabe JWR
Nat Commun (2020) 11 pp. 3252-3252 [ Pubmed: 32591534 DOI: doi:10.1038/s41467-020-17078-8 ]
- Zinc ion (65 Da, Ligand)
- Potassium ion (39 Da, Ligand)
- Deoxynucleotidyltransferase terminal-interacting protein 1 (14 kDa, Protein from Homo sapiens)
- Inositol hexakisphosphate (660 Da, Ligand)
- Dimeric complex of the midac deacetylase complex containing hdac1, the elm2-sant domain of mideas and the dimerisation domain of dnttip1 (178 kDa, Complex from Homo sapiens)
- Histone deacetylase 1 (55 kDa, Protein from Homo sapiens)
- Mitotic deacetylase-associated sant domain protein (19 kDa, Protein from Homo sapiens)
The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 6z2k
Q-score: 0.206
Turnbull RE, Fairall L, Saleh A, Kelsall E, Morris KL, Ragan TJ, Savva CG, Chandru A, Millard CJ, Makarova OV, Smith CJ, Roseman AM, Fry AM, Cowley SM, Schwabe JWR
Nat Commun (2020) 11 pp. 3252-3252 [ Pubmed: 32591534 DOI: doi:10.1038/s41467-020-17078-8 ]
- Zinc ion (65 Da, Ligand)
- Potassium ion (39 Da, Ligand)
- Tetrameric complex of the midac deacetylase complex containing hdac1, the elm2-sant domain of mideas and the dimerisation domain of dnttip1 (357 kDa, Complex from Homo sapiens)
- Deoxynucleotidyltransferase terminal-interacting protein 1 (14 kDa, Protein from Homo sapiens)
- Inositol hexakisphosphate (660 Da, Ligand)
- Histone deacetylase 1 (55 kDa, Protein from Homo sapiens)
- Mitotic deacetylase-associated sant domain protein (19 kDa, Protein from Homo sapiens)
Negative stain map of CoREST complex (LSD1:RCOR1:HDAC1)
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
Cell Rep (2020) 30 pp. 2699-2711.e8 [ DOI: doi:10.1016/j.celrep.2020.01.091 Pubmed: 32101746 ]
Cryo-EM map of glutaraldehye cross-linked CoREST complex (LSD1:RCOR1:HDAC1)
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
Cell Rep (2020) 30 pp. 2699-2711.e8 [ DOI: doi:10.1016/j.celrep.2020.01.091 Pubmed: 32101746 ]
Cryo EM map of BS3 crosslinked CoREST complex (LSD1:RCOR1:HDAC1)- closed form
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
Cell Rep (2020) 30 pp. 2699-2711.e8 [ DOI: doi:10.1016/j.celrep.2020.01.091 Pubmed: 32101746 ]
Cryo EM map of BS3 crosslinked CoREST complex (LSD1:RCOR1:HDAC1) - open form
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
Cell Rep (2020) 30 pp. 2699-2711.e8 [ DOI: doi:10.1016/j.celrep.2020.01.091 Pubmed: 32101746 ]
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
Cell Rep (2020) 30 pp. 2699-2711.e8 [ DOI: doi:10.1016/j.celrep.2020.01.091 Pubmed: 32101746 ]
- Nucleosome with corest complex (rcor1: lsd1:hdac1) (410 kDa, Complex from Homo sapiens)