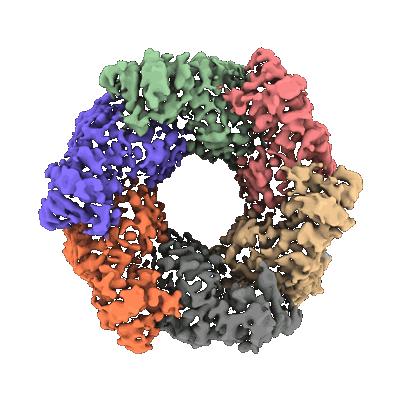
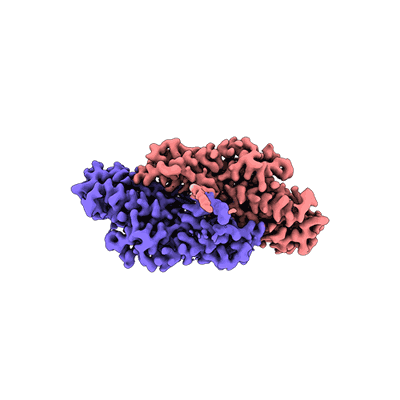
Progesterone-bound DB3 Fab in complex with computationally designed DBPro1156_2 protein binder
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 9fkd
Q-score: 0.488
Marchand A, Buckley S, Schneuing A, Pacesa M, Elia M, Gainza P, Elizarova E, Neeser RM, Lee PW, Reymond L, Miao Y, Scheller L, Georgeon S, Schmidt J, Schwaller P, Maerkl SJ, Bronstein M, Correia BE
Nature (2025) [ Pubmed: 39814890 DOI: doi:10.1038/s41586-024-08435-4 ]
- Db3 fab light chain (24 kDa, Protein from synthetic construct)
- Progesterone-bound db3 fab in complex with computationally designed dbpro1156_2 protein binder (107 kDa, Complex from synthetic construct)
- Progesterone (314 Da, Ligand)
- Anti-kappa fab light chain (24 kDa, Protein from synthetic construct)
- Anti-kappa fab heavy chain (25 kDa, Protein from synthetic construct)
- De novo designed dbpro1156_2 binder (8 kDa, Protein from synthetic construct)
- Db3 fab heavy chain (25 kDa, Protein from synthetic construct)
Resolution: 4.16 Å
EM Method: Single-particle
Fitted PDBs: 9bei
Q-score: 0.276
Goverde CA, Pacesa M, Goldbach N, Dornfeld LJ, Balbi PEM, Georgeon S, Rosset S, Kapoor S, Choudhury J, Dauparas J, Schellhaas C, Kozlov S, Baker D, Ovchinnikov S, Vecchio AJ, Correia BE
Nature (2024) 631 pp. 449-458 [ Pubmed: 38898281 DOI: doi:10.1038/s41586-024-07601-y ]
- Heat-labile enterotoxin b chain (14 kDa, Protein from Clostridium perfringens)
- Anti-fab nanobody (13 kDa, Protein from Escherichia coli)
- Cop-2 fab heavy chain (25 kDa, Protein from Escherichia coli)
- Cop-2 fab light chain (23 kDa, Protein from Escherichia coli)
- Claudin-4 (22 kDa, Protein from Homo sapiens)
- Synthetic human claudin-4 complex with clostridium perfringens enterotoxin c-terminal domain, sfab cop-2, and nanobody against cop-2 (103 kDa, Complex from Homo sapiens)
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus
Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 8p49
Q-score: 0.504
Schweke H, Pacesa M, Levin T, Goverde CA, Kumar P, Duhoo Y, Dornfeld LJ, Dubreuil B, Georgeon S, Ovchinnikov S, Woolfson DN, Correia BE, Dey S, Levy ED
Cell (2024) 187 pp. 999- [ Pubmed: 38325366 DOI: doi:10.1016/j.cell.2024.01.022 ]
Cysteine tRNA ligase homodimer
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8qhp
Q-score: 0.546
Schweke H, Pacesa M, Levin T, Goverde CA, Kumar P, Duhoo Y, Dornfeld LJ, Dubreuil B, Georgeon S, Ovchinnikov S, Woolfson DN, Correia BE, Dey S, Levy ED
Cell (2024) 187 pp. 999- [ Pubmed: 38325366 DOI: doi:10.1016/j.cell.2024.01.022 ]
- Cysteine--trna ligase (57 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Homodimeric form of cysteine trna ligase (114 kDa, Complex from Pyrococcus furiosus DSM 3638)
- Zinc ion (65 Da, Ligand)
cryo-EM structure of omicron spike in complex with de novo designed binder, full map
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7zrv
Q-score: 0.4
Gainza P, Wehrle S, Van Hall-Beauvais A, Marchand A, Scheck A, Harteveld Z, Buckley S, Ni D, Tan S, Sverrisson F, Goverde C, Turelli P, Raclot C, Teslenko A, Pacesa M, Rosset S, Georgeon S, Marsden J, Petruzzella A, Liu K, Xu Z, Chai Y, Han P, Gao GF, Oricchio E, Fierz B, Trono D, Stahlberg H, Bronstein M, Correia BE
Nature (2023) 617 pp. 176-184 [ DOI: doi:10.1038/s41586-023-05993-x Pubmed: 37100904 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Omicron spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- De novo designed binder (8 kDa, Protein from Drosophila melanogaster)
- Omicron spike in complex with de novo designed binder, full map (Complex)
- De novo designed binder (Complex from Drosophila melanogaster)
- Spike glycoprotein,envelope glycoprotein (142 kDa, Protein from Tequatrovirus T4)
cryo-EM structure of omicron spike in complex with de novo designed binder, local
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 7zsd
Q-score: 0.431
Gainza P, Wehrle S, Van Hall-Beauvais A, Marchand A, Scheck A, Harteveld Z, Buckley S, Ni D, Tan S, Sverrisson F, Goverde C, Turelli P, Raclot C, Teslenko A, Pacesa M, Rosset S, Georgeon S, Marsden J, Petruzzella A, Liu K, Xu Z, Chai Y, Han P, Gao GF, Oricchio E, Fierz B, Trono D, Stahlberg H, Bronstein M, Correia BE
Nature (2023) 617 pp. 176-184 [ DOI: doi:10.1038/s41586-023-05993-x Pubmed: 37100904 ]
- Spike glycoprotein (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Omicron spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- De novo designed binder (8 kDa, Protein from Drosophila melanogaster)
- Omicron spike in complex with de novo designed binder (450 kDa, Complex)
- De novo designed binder (Complex from Drosophila melanogaster)
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 7zss
Q-score: 0.453
Gainza P, Wehrle S, Van Hall-Beauvais A, Marchand A, Scheck A, Harteveld Z, Buckley S, Ni D, Tan S, Sverrisson F, Goverde C, Turelli P, Raclot C, Teslenko A, Pacesa M, Rosset S, Georgeon S, Marsden J, Petruzzella A, Liu K, Xu Z, Chai Y, Han P, Gao GF, Oricchio E, Fierz B, Trono D, Stahlberg H, Bronstein M, Correia BE
Nature (2023) 617 pp. 176-184 [ DOI: doi:10.1038/s41586-023-05993-x Pubmed: 37100904 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- D614g spike in complex with de novo designed binder (450 kDa, Complex)
- De novo designed binder (8 kDa, Protein from Drosophila melanogaster)
- De novo designed binder (Complex from Drosophila melanogaster)
- Spike glycoprotein (126 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- D614g spike (Complex from Severe acute respiratory syndrome coronavirus 2)