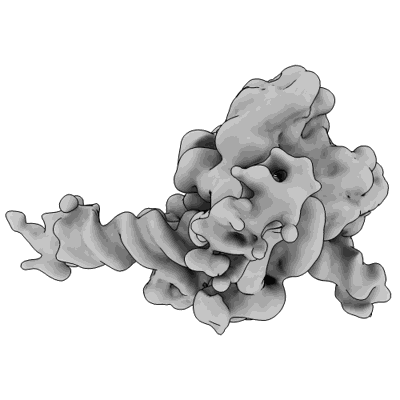
CryoEM structure of canonical rice nucleosome core particle
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8q15
Q-score: 0.401
Hari Sundar G V, Sotelo-Parrilla P, Raju S, Jha S, Gireesh A, Gut F, Vinothkumar KR, Berger F, Jeyaprakash AA, Shivaprasad PV
To Be Published
- Histone h2b.4 (16 kDa, Protein from Oryza)
- Histone h2a.2 (13 kDa, Protein from Oryza)
- Rice canonical nucleosome core particle with widom 601 dna sequence (Complex from Oryza)
- Histone h4 (11 kDa, Protein from Oryza)
- Widom 601 (44 kDa, DNA from synthetic construct)
- Widom 601 (45 kDa, DNA from synthetic construct)
- Histone h3.2 (15 kDa, Protein from Oryza)
CryoEM structure of rice nucleosome containing a H4 variant chimera
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8q16
Q-score: 0.441
Hari Sundar G V, Sotelo-Parrilla P, Raju S, Jha S, Gireesh A, Gut F, Vinothkumar KR, Berger F, Jeyaprakash AA, Shivaprasad PV
To Be Published
- Histone h3.2 (15 kDa, Protein from Oryza)
- Widom 601 (45 kDa, DNA from synthetic construct)
- Chimeric h4.v (11 kDa, Protein from Oryza)
- Histone h2a.2 (13 kDa, Protein from Oryza)
- Widom 601 (45 kDa, DNA from synthetic construct)
- Rice nucleosome core particle containing a histone h4 variant chimera with widom 601 dna (Complex from Oryza)
- Histone h2b.4 (16 kDa, Protein from Oryza)
Human OGG1 bound to a nucleosome core particle with 8-oxodGuo lesion at SHL6.0
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 9eoz
Q-score: 0.465
Ren M, Gut F, Fan Y, Ma J, Shan X, Yikilmazsoy A, Likhodeeva M, Hopfner KP, Zhou C
Nat Commun (2024) 15 pp. 9407-9407 [ Pubmed: 39477986 DOI: doi:10.1038/s41467-024-53811-3 ]
- Histone h2b type 1-c/e/f/g/i (13 kDa, Protein from Homo sapiens)
- Widom 601 dna (145-mer) (45 kDa, DNA from synthetic construct)
- Widom 601 dna (Complex from synthetic construct)
- Histone h4 (11 kDa, Protein from Homo sapiens)
- Binary complex of human 8-oxoguanine glycosylase with nucleosome core particle (Complex)
- Widom 601 dna (145-mer) (44 kDa, DNA from synthetic construct)
- Histone h3.3 (15 kDa, Protein from Homo sapiens)
- N-glycosylase/dna lyase (40 kDa, Protein from Homo sapiens)
- Nucleosome core particle (Complex from Homo sapiens)
- Histone h2a type 1-c (14 kDa, Protein from Homo sapiens)
Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7yzo
Q-score: 0.491
Gut F, Kashammer L, Lammens K, Bartho JD, Boggusch AM, van de Logt E, Kessler B, Hopfner KP
Mol Cell (2022) 82 pp. 3513-3522.e6 [ Pubmed: 35987200 DOI: doi:10.1016/j.molcel.2022.07.019 ]
- Dna (31-mer) (36 kDa, DNA from synthetic construct)
- Nuclease sbccd subunit c (118 kDa, Protein from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Dna (31-mer) (37 kDa, DNA from synthetic construct)
- Nuclease sbccd subunit d (45 kDa, Protein from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Complex of mre11 dimer, rad50 dimer and double-stranded dna (Complex from Escherichia coli)
- Manganese (ii) ion (54 Da, Ligand)
Gut F, Kashammer L, Lammens K, Bartho JD, Boggusch AM, van de Logt E, Kessler B, Hopfner KP
Mol Cell (2022) 82 pp. 3513-3522.e6 [ Pubmed: 35987200 DOI: doi:10.1016/j.molcel.2022.07.019 ]
Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 7yzp
Q-score: 0.394
Gut F, Kashammer L, Lammens K, Bartho JD, Boggusch AM, van de Logt E, Kessler B, Hopfner KP
Mol Cell (2022) 82 pp. 3513-3522.e6 [ Pubmed: 35987200 DOI: doi:10.1016/j.molcel.2022.07.019 ]
- Nuclease sbccd subunit c (118 kDa, Protein from Escherichia coli)
- Dna hairpin (Complex from DNA molecule)
- Complex of mre11 dimer, rad50 dimer and a dna hairpin (Complex)
- Magnesium ion (24 Da, Ligand)
- Nuclease sbccd (Complex from Escherichia coli)
- Nuclease sbccd subunit d (45 kDa, Protein from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna hairpin (59-mer) (23 kDa, DNA from synthetic construct)
- Manganese (ii) ion (54 Da, Ligand)
C. thermophilum Ku70/80 heterodimer bound to DNA
Gut F, Kashammer L, Lammens K, Bartho JD, Boggusch AM, van de Logt E, Kessler B, Hopfner KP
Mol Cell (2022) 82 pp. 3513-3522.e6 [ DOI: doi:10.1016/j.molcel.2022.07.019 Pubmed: 35987200 ]
Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 7z03
Q-score: 0.253
Gut F, Kashammer L, Lammens K, Bartho JD, Boggusch AM, van de Logt E, Kessler B, Hopfner KP
Mol Cell (2022) 82 pp. 3513-3522.e6 [ Pubmed: 35987200 DOI: doi:10.1016/j.molcel.2022.07.019 ]
- Dna (39-mer) (36 kDa, DNA from synthetic construct)
- Nuclease sbccd subunit c (118 kDa, Protein from Escherichia coli)
- Dna (39-mer) (37 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Nuclease sbccd subunit d (45 kDa, Protein from Escherichia coli)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Complex of mre11 dimer, rad50 dimer and double-stranded dna (Complex from Escherichia coli)
- Manganese (ii) ion (54 Da, Ligand)
Resting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ATPgS
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 6s6v
Q-score: 0.486
Kashammer L, Saathoff JH, Lammens K, Gut F, Bartho J, Alt A, Kessler B, Hopfner KP
Mol Cell (2019) 76 pp. 382- [ DOI: doi:10.1016/j.molcel.2019.07.035 Pubmed: 31492634 ]
- Nuclease sbccd subunit d (45 kDa, Protein from Escherichia coli)
- Heterotetrameric complex of full-length mre11-rad50 in complex with atpgs (Complex from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Manganese (ii) ion (54 Da, Ligand)
- Nuclease sbccd subunit c (118 kDa, Protein from Escherichia coli)
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
Kashammer L, Saathoff JH, Lammens K, Gut F, Bartho J, Alt A, Kessler B, Hopfner KP
Mol Cell (2019) 76 pp. 382-394.e6 [ Pubmed: 31492634 DOI: doi:10.1016/j.molcel.2019.07.035 ]