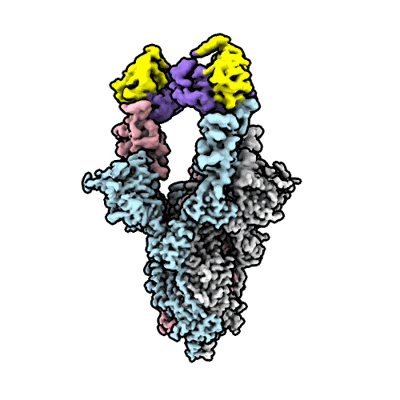
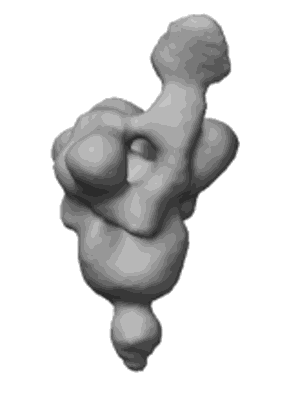Du W, Janssens R, Mykytyn AZ, Li W, Drabek D, van Haperen R, Chatziandreou M, Rissmann M, van der Lee J, van Dortmondt M, Martin IS, van Kuppeveld FJM, Hurdiss DL, Haagmans BL, Grosveld F, Bosch BJ
Front Immunol (2023) 14 pp. 1111385-1111385 [ DOI: doi:10.3389/fimmu.2023.1111385 Pubmed: 36895554 ]
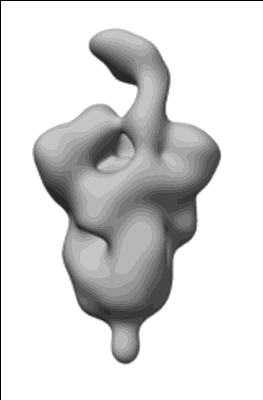
Du W, Janssens R, Mykytyn AZ, Li W, Drabek D, van Haperen R, Chatziandreou M, Rissmann M, van der Lee J, van Dortmondt M, Martin IS, van Kuppeveld FJM, Hurdiss DL, Haagmans BL, Grosveld F, Bosch BJ
Front Immunol (2023) 14 pp. 1111385-1111385 [ DOI: doi:10.3389/fimmu.2023.1111385 Pubmed: 36895554 ]
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 8c8p
Q-score: 0.22
Du W, Janssens R, Mykytyn AZ, Li W, Drabek D, van Haperen R, Chatziandreou M, Rissmann M, van der Lee J, van Dortmondt M, Martin IS, van Kuppeveld FJM, Hurdiss DL, Haagmans BL, Grosveld F, Bosch BJ
Front Immunol (2023) 14 pp. 1111385-1111385 [ DOI: doi:10.3389/fimmu.2023.1111385 Pubmed: 36895554 ]
- Heavy-chain-only antibody 10d12 (41 kDa, Protein from Mus musculus)
- Spike glycoprotein,sars-cov-2 spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike glycoprotein in complex with the 10d12 heavy-chain-only antibody (Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 7r40
Q-score: 0.333
Du W, Hurdiss DL, Drabek D, Mykytyn AZ, Kaiser FK, Gonzalez-Hernandez M, Munoz-Santos D, Lamers MM, van Haperen R, Li W, Drulyte I, Wang C, Sola I, Armando F, Beythien G, Ciurkiewicz M, Baumgartner W, Guilfoyle K, Smits T, van der Lee J, van Kuppeveld FJM, van Amerongen G, Haagmans BL, Enjuanes L, Osterhaus ADME, Grosveld F, Bosch BJ
Sci Immunol (2022) 7 pp. eabp9312-eabp9312 [ DOI: doi:10.1126/sciimmunol.abp9312 Pubmed: 35471062 ]
- 87g7 light chain variable region (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (141 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 87g7 antibody fab fragment (Complex from Homo sapiens)
- Sars-cov-2 spike glycoprotein in complex with the 87g7 antibody fab fragment (500 kDa, Complex)
- 87g7 heavy chain variable region (23 kDa, Protein from Homo sapiens)
Du W, Hurdiss DL, Drabek D, Mykytyn AZ, Kaiser FK, Gonzalez-Hernandez M, Munoz-Santos D, Lamers MM, van Haperen R, Li W, Drulyte I, Wang C, Sola I, Armando F, Beythien G, Ciurkiewicz M, Baumgartner W, Guilfoyle K, Smits T, van der Lee J, van Kuppeveld FJM, van Amerongen G, Haagmans BL, Enjuanes L, Osterhaus ADME, Grosveld F, Bosch BJ
Sci Immunol (2022) 7 pp. eabp9312-eabp9312 [ DOI: doi:10.1126/sciimmunol.abp9312 Pubmed: 35471062 ]
- Sars-cov-2 spike protein (Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab (Complex from Homo sapiens)
- 87g7 light chain variable domain (Protein from Homo sapiens)
- Sars-cov-2 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- 87g7 heavy chain variable domain (Protein from Homo sapiens)
- Sars-cov-2 spike glycoprotein in complex with the 87g7 antibody fab fragment (500 kDa, Complex)
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-15 Fab
Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ
Science (2020) 369 pp. 643-650 [ DOI: doi:10.1126/science.abc5902 Pubmed: 32540902 ]
negative stain EM map of SARS-CoV-2 spike in complex with COVA1-22
Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ
Science (2020) 369 pp. 643-650 [ DOI: doi:10.1126/science.abc5902 Pubmed: 32540902 ]
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-07
Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ
Science (2020) 369 pp. 643-650 [ DOI: doi:10.1126/science.abc5902 Pubmed: 32540902 ]
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-39 Fab
Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ
Science (2020) 369 pp. 643-650 [ DOI: doi:10.1126/science.abc5902 Pubmed: 32540902 ]
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-04 Fab
Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ
Science (2020) 369 pp. 643-650 [ DOI: doi:10.1126/science.abc5902 Pubmed: 32540902 ]