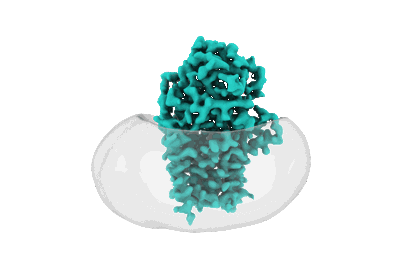
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8b0k
Q-score: 0.541
Smithers L, Degtjarik O, Weichert D, Huang CY, Boland C, Bowen K, Oluwole A, Lutomski C, Robinson CV, Scanlan EM, Wang M, Olieric V, Shalev-Benami M, Caffrey M
Sci Adv (2023) 9 pp. eadf5799-eadf5799 [ Pubmed: 37390210 DOI: doi:10.1126/sciadv.adf5799 ]
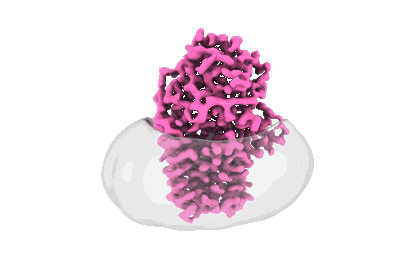
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 8b0l
Q-score: 0.511
Smithers L, Degtjarik O, Weichert D, Huang CY, Boland C, Bowen K, Oluwole A, Lutomski C, Robinson CV, Scanlan EM, Wang M, Olieric V, Shalev-Benami M, Caffrey M
Sci Adv (2023) 9 pp. eadf5799-eadf5799 [ Pubmed: 37390210 DOI: doi:10.1126/sciadv.adf5799 ]
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 8b0m
Q-score: 0.552
Smithers L, Degtjarik O, Weichert D, Huang CY, Boland C, Bowen K, Oluwole A, Lutomski C, Robinson CV, Scanlan EM, Wang M, Olieric V, Shalev-Benami M, Caffrey M
Sci Adv (2023) 9 pp. eadf5799-eadf5799 [ Pubmed: 37390210 DOI: doi:10.1126/sciadv.adf5799 ]
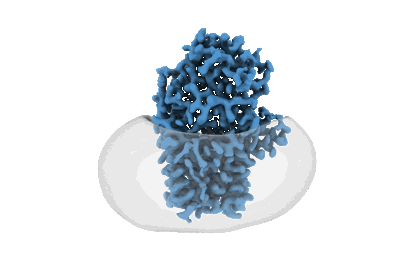
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE
Resolution: 2.67 Å
EM Method: Single-particle
Fitted PDBs: 8b0n
Q-score: 0.6
Smithers L, Degtjarik O, Weichert D, Huang CY, Boland C, Bowen K, Oluwole A, Lutomski C, Robinson CV, Scanlan EM, Wang M, Olieric V, Shalev-Benami M, Caffrey M
Sci Adv (2023) 9 pp. eadf5799-eadf5799 [ Pubmed: 37390210 DOI: doi:10.1126/sciadv.adf5799 ]
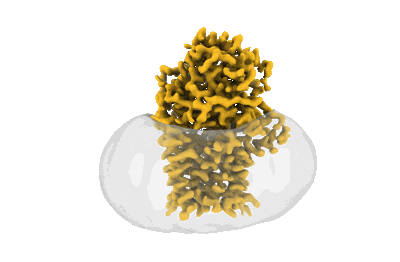
Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3
Resolution: 3.02 Å
EM Method: Single-particle
Fitted PDBs: 8b0o
Q-score: 0.534
Smithers L, Degtjarik O, Weichert D, Huang CY, Boland C, Bowen K, Oluwole A, Lutomski C, Robinson CV, Scanlan EM, Wang M, Olieric V, Shalev-Benami M, Caffrey M
Sci Adv (2023) 9 pp. eadf5799-eadf5799 [ Pubmed: 37390210 DOI: doi:10.1126/sciadv.adf5799 ]
- [(2~{r})-3-[(2~{r})-3-[[(2~{r})-1-[[(2~{r})-1-[[(2~{r})-6-[(2-aminophenyl)carbonylamino]-1-azanyl-1-oxidanylidene-hexan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate (1 kDa, Ligand)
- Apolipoprotein n-acyltransferase (Cellular component from Escherichia coli K-12)
- Apolipoprotein n-acyltransferase (59 kDa, Protein from Escherichia coli K-12)
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3
Resolution: 2.86 Å
EM Method: Single-particle
Fitted PDBs: 8b0p
Q-score: 0.564
Smithers L, Degtjarik O, Weichert D, Huang CY, Boland C, Bowen K, Oluwole A, Lutomski C, Robinson CV, Scanlan EM, Wang M, Olieric V, Shalev-Benami M, Caffrey M
Sci Adv (2023) 9 pp. eadf5799-eadf5799 [ Pubmed: 37390210 DOI: doi:10.1126/sciadv.adf5799 ]
- Pam3-skkkk (621 Da, Protein from synthetic construct)
- Apolipoprotein n-acyltransferase (59 kDa, Protein from Escherichia coli K-12)
- Apolipoprotein n-acyltransferase (Cellular component from Escherichia coli K-12)
- [(2~{s})-3-[(2~{s})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate (909 Da, Ligand)
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 7w6m
Q-score: 0.267