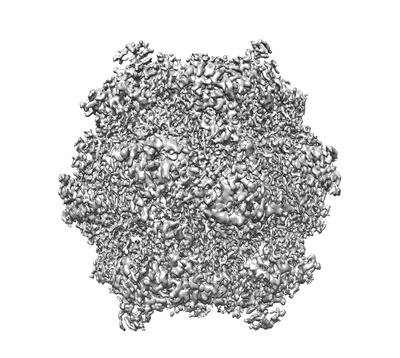
CryoEM structure of the Dsl1 complex bound to SNAREs Sec20 and Use1
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 8eki
Q-score: 0.182
DAmico KA, Stanton AE, Shirkey JD, Travis SM, Jeffrey PD, Hughson FM
Nat Struct Mol Biol (2024) 31 pp. 246-254 [ DOI: doi:10.1038/s41594-023-01164-8 Pubmed: 38196032 ]
- Protein transport protein tip20 (81 kDa, Protein from Saccharomyces cerevisiae S288C)
- Protein transport protein use1 (24 kDa, Protein from Saccharomyces cerevisiae S288C)
- Dsl1 complex (Complex from Saccharomyces cerevisiae)
- Dsl1 complex bound to snare proteins sec20 and use1 (255 kDa, Complex from Saccharomyces cerevisiae)
- Protein transport protein sec20 (32 kDa, Protein from Saccharomyces cerevisiae S288C)
- Protein transport protein dsl1 (91 kDa, Protein from Saccharomyces cerevisiae S288C)
- Protein transport protein sec39 (82 kDa, Protein from Saccharomyces cerevisiae S288C)
Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18
Resolution: 5.1 Å
EM Method: Single-particle
Fitted PDBs: 8dit
Q-score: 0.105
Port SA, Farrell PD, Jeffrey PD, DiMaio F, Hughson FM
To Be Published
- Vacuolar protein sorting-associated protein 33 (77 kDa, Protein from Chaetomium thermophilum)
- Hops core complex (171 kDa, Complex from Chaetomium thermophilum)
- Vps18 (Complex from Chaetomium thermophilum)
- His-vps33:vps16 (Complex from Chaetomium thermophilum)
- Vacuolar protein sorting-associated protein 16 (94 kDa, Protein from Chaetomium thermophilum)
- Vacuolar protein sorting-associated protein 18 (108 kDa, Protein from Chaetomium thermophilum)
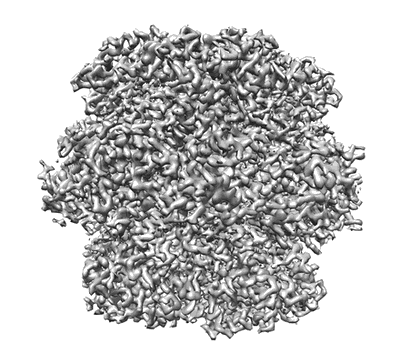
Resolution: 2.13 Å
EM Method: Single-particle
Fitted PDBs: 7jfo
Q-score: 0.734
He S, Chou HT, Matthies D, Wunder T, Meyer MT, Atkinson N, Martinez-Sanchez A, Jeffrey PD, Port SA, Patena W, He G, Chen VK, Hughson FM, McCormick AJ, Mueller-Cajar O, Engel BD, Yu Z, Jonikas MC
Nat Plants (2020) 6 pp. 1480-1490 [ DOI: doi:10.1038/s41477-020-00811-y Pubmed: 33230314 ]
- Water (18 Da, Ligand)
- Lci5 (2 kDa, Protein from Chlamydomonas reinhardtii)
- Epyc1(49-72) peptide-bound rubisco (Complex from Chlamydomonas reinhardtii)
- Ribulose bisphosphate carboxylase large chain (52 kDa, Protein from Chlamydomonas reinhardtii)
- Ribulose bisphosphate carboxylase small chain 2, chloroplastic (20 kDa, Protein from Chlamydomonas reinhardtii)
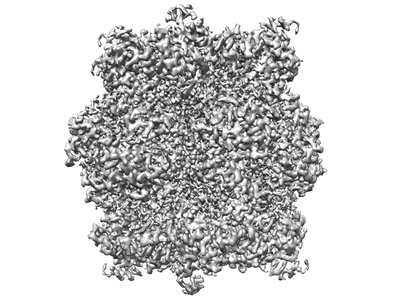
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 7jn4
Q-score: 0.624
He S, Chou HT, Matthies D, Wunder T, Meyer MT, Atkinson N, Martinez-Sanchez A, Jeffrey PD, Port SA, Patena W, He G, Chen VK, Hughson FM, McCormick AJ, Mueller-Cajar O, Engel BD, Yu Z, Jonikas MC
Nat Plants (2020) 6 pp. 1480-1490 [ DOI: doi:10.1038/s41477-020-00811-y Pubmed: 33230314 ]
- Ribulose bisphosphate carboxylase large chain (52 kDa, Protein from Chlamydomonas reinhardtii)
- Water (18 Da, Ligand)
- Rubisco at apo state (Complex from Chlamydomonas reinhardtii)
- Ribulose bisphosphate carboxylase small chain 2, chloroplastic (20 kDa, Protein from Chlamydomonas reinhardtii)
EPYC1(106-135) peptide-bound Rubisco
Resolution: 2.06 Å
EM Method: Single-particle
Fitted PDBs: 7jsx
Q-score: 0.677
He S, Chou HT, Matthies D, Wunder T, Meyer MT, Atkinson N, Martinez-Sanchez A, Jeffrey PD, Port SA, Patena W, He G, Chen VK, Hughson FM, McCormick AJ, Mueller-Cajar O, Engel BD, Yu Z, Jonikas MC
Nat Plants (2020) 6 pp. 1480-1490 [ DOI: doi:10.1038/s41477-020-00811-y Pubmed: 33230314 ]
- Water (18 Da, Ligand)
- Ribulose bisphosphate carboxylase large chain (52 kDa, Protein from Chlamydomonas reinhardtii)
- Ribulose bisphosphate carboxylase small chain 2, chloroplastic (20 kDa, Protein from Chlamydomonas reinhardtii)
- Epyc1(106-135) peptide-bound rubisco (Complex from Chlamydomonas reinhardtii)
- Epyc1 (3 kDa, Protein from Chlamydomonas reinhardtii)