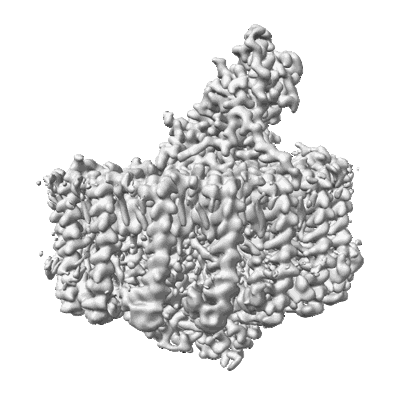
Resolution: 2.24 Å
EM Method: Single-particle
Fitted PDBs: 8wdu
Q-score: 0.676
Tani K, Kanno R, Harada A, Kobayashi Y, Minamino A, Takenaka S, Nakamura N, Ji XC, Purba ER, Hall M, Yu LJ, Madigan MT, Mizoguchi A, Iwasaki K, Humbel BM, Kimura Y, Wang-Otomo ZY
Commun Biol (2024) 7 pp. 176-176 [ Pubmed: 38347078 DOI: doi:10.1038/s42003-024-05863-w ]
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- (1r)-2-{[{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl (11e)-octadec-11-enoate (749 Da, Ligand)
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Bacteriochlorophyll a (911 Da, Ligand)
- Reaction center protein l chain (31 kDa, Protein from Allochromatium vinosum DSM 180)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- (2s)-3-hydroxypropane-1,2-diyl dihexadecanoate (568 Da, Ligand)
- Reaction center protein m chain (36 kDa, Protein from Allochromatium vinosum DSM 180)
- Ubiquinone-8 (727 Da, Ligand)
- Photosynthetic lh1-rc complex from the purple sulfur phototrophic bacterium allochromatium vinosum (Complex from Allochromatium vinosum DSM 180)
- Menaquinone 8 (717 Da, Ligand)
- Photosynthetic reaction center cytochrome c subunit (41 kDa, Protein from Allochromatium vinosum DSM 180)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Water (18 Da, Ligand)
- Lauryl dimethylamine-n-oxide (229 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Cardiolipin (1 kDa, Ligand)
- Magnesium ion (24 Da, Ligand)
- Photosynthetic reaction center h subunit (28 kDa, Protein from Allochromatium vinosum DSM 180)
- Spirilloxanthin (596 Da, Ligand)
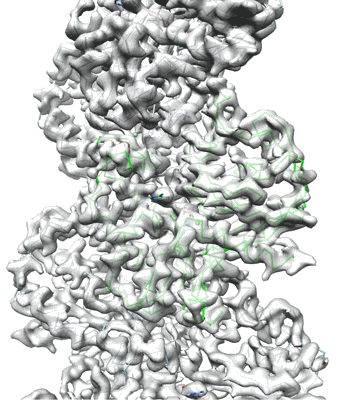
Resolution: 2.24 Å
EM Method: Single-particle
Fitted PDBs: 8wdv
Q-score: 0.658
Tani K, Kanno R, Harada A, Kobayashi Y, Minamino A, Takenaka S, Nakamura N, Ji XC, Purba ER, Hall M, Yu LJ, Madigan MT, Mizoguchi A, Iwasaki K, Humbel BM, Kimura Y, Wang-Otomo ZY
Commun Biol (2024) 7 pp. 176-176 [ DOI: doi:10.1038/s42003-024-05863-w Pubmed: 38347078 ]
- Reaction center protein m chain (36 kDa, Protein from Allochromatium vinosum DSM 180)
- (1r)-2-{[{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl (11e)-octadec-11-enoate (749 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Calcium ion (40 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- Cardiolipin (1 kDa, Ligand)
- Lauryl dimethylamine-n-oxide (229 Da, Ligand)
- Spirilloxanthin (596 Da, Ligand)
- Photosynthetic reaction center cytochrome c subunit (41 kDa, Protein from Allochromatium vinosum DSM 180)
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Photosynthetic lh1-rc complex from the purple sulfur phototrophic bacterium allochromatium vinosum (Complex from Allochromatium vinosum DSM 180)
- Reaction center protein l chain (31 kDa, Protein from Allochromatium vinosum DSM 180)
- Ubiquinone-8 (727 Da, Ligand)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- (2s)-3-hydroxypropane-1,2-diyl dihexadecanoate (568 Da, Ligand)
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Water (18 Da, Ligand)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Photosynthetic reaction center h subunit (28 kDa, Protein from Allochromatium vinosum DSM 180)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Menaquinone 8 (717 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Bacteriochlorophyll a (911 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
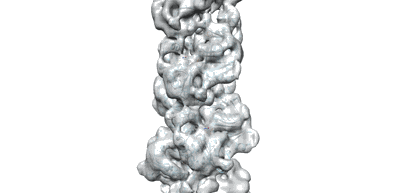
Resolution: 3.9 Å
EM Method: Helical reconstruction
Fitted PDBs: 7x54
Q-score: 0.375
Koh A, Ali S, Popp D, Tanaka K, Kitaoku Y, Miyazaki N, Iwasaki K, Mitsuoka K, Robert R, Narita A
To Be Published
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Cbg-parm filament with adp (Complex from Clostridium botulinum)
- Parm/stba family protein (32 kDa, Protein from Clostridium botulinum Bf)
A CBg-ParM filament with GTP and a short incubation time
Resolution: 8.6 Å
EM Method: Helical reconstruction
Fitted PDBs: 7x55
Q-score: 0.168
Koh A, Ali S, Popp D, Tanaka K, Kitaoku Y, Miyazaki N, Iwasaki K, Mitsuoka K, Robert R, Narita A
To Be Published
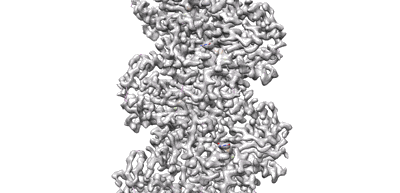
Resolution: 3.5 Å
EM Method: Helical reconstruction
Fitted PDBs: 7x56
Q-score: 0.486
Koh A, Ali S, Popp D, Tanaka K, Kitaoku Y, Miyazaki N, Iwasaki K, Mitsuoka K, Robert R, Narita A
To Be Published
- Cbg-parm filament with gdp (Complex from Clostridium botulinum)
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Parm/stba family protein (32 kDa, Protein from Clostridium botulinum Bf)
A CBg-ParM filament with GTP or GDPPi
Resolution: 6.5 Å
EM Method: Helical reconstruction
Fitted PDBs: 7x59
Q-score: 0.189
Koh A, Ali S, Popp D, Tanaka K, Kitaoku Y, Miyazaki N, Iwasaki K, Mitsuoka K, Robert R, Narita A
To Be Published
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Cbg-parm filament with gtp or gdppi (Complex from Clostridium botulinum)
- Parm/stba family protein (32 kDa, Protein from Clostridium botulinum)
Cryo-EM structure of phosphoketolase from Bifidobacterium longum
Resolution: 2.1 Å
EM Method: Single-particle
Fitted PDBs: 6lxv
Q-score: 0.763
Nakata K, Miyazaki N, Yamaguchi H, Hirose M, Kashiwagi T, Kutumbarao NHV, Miyashita O, Tama F, Miyano H, Mizukoshi T, Iwasaki K
J Struct Biol (2022) 214 pp. 107842-107842 [ Pubmed: 35181457 DOI: doi:10.1016/j.jsb.2022.107842 ]
- Water (18 Da, Ligand)
- Phosphoketolase (93 kDa, Protein from Bifidobacterium longum subsp. longum F8)
- Phosphoketolase with thiamine-diphophate (Complex from Bifidobacterium longum subsp. longum F8)
- Calcium ion (40 Da, Ligand)
- Thiamine diphosphate (425 Da, Ligand)
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 6klo
Q-score: 0.586
Yamada T, Yoshida T, Kawamoto A, Mitsuoka K, Iwasaki K, Tsuge H
Nat Struct Mol Biol (2020) 27 pp. 288-296 [ DOI: doi:10.1038/s41594-020-0388-6 Pubmed: 32123390 ]
- Calcium ion (40 Da, Ligand)
- Iota toxin component ib (74 kDa, Protein from Clostridium perfringens)
- Complex structure of iota toxin enzymatic component (ia) and binding component (ib) pore with short stem (570 kDa, Complex from Clostridium perfringens)
- Iota toxin component ia (48 kDa, Protein from Clostridium perfringens)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6klw
Q-score: 0.572
Yamada T, Yoshida T, Kawamoto A, Mitsuoka K, Iwasaki K, Tsuge H
Nat Struct Mol Biol (2020) 27 pp. 288-296 [ Pubmed: 32123390 DOI: doi:10.1038/s41594-020-0388-6 ]
- Complex structure of iota toxin enzymatic component (ia) and binding component (ib) pore with long stem (570 kDa, Complex from Clostridium perfringens)
- Calcium ion (40 Da, Ligand)
- Iota toxin component ib (74 kDa, Protein from Clostridium perfringens)
- Iota toxin component ia (48 kDa, Protein from Clostridium perfringens)
Pore structure of Iota toxin binding component (Ib)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6klx
Q-score: 0.596
Yamada T, Yoshida T, Kawamoto A, Mitsuoka K, Iwasaki K, Tsuge H
Nat Struct Mol Biol (2020) 27 pp. 288-296 [ DOI: doi:10.1038/s41594-020-0388-6 Pubmed: 32123390 ]
- Calcium ion (40 Da, Ligand)
- Iota toxin component ib (74 kDa, Protein from Clostridium perfringens)
- Pore structure of iota toxin binding component (ib) (520 kDa, Complex from Clostridium perfringens)