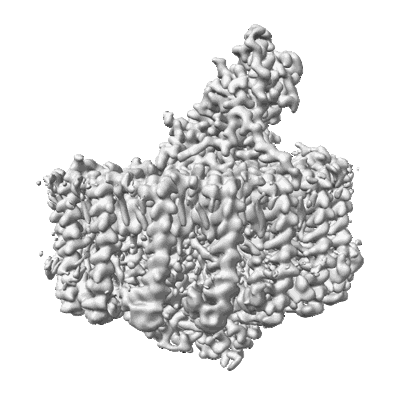
Resolution: 2.24 Å
EM Method: Single-particle
Fitted PDBs: 8wdu
Q-score: 0.676
Tani K, Kanno R, Harada A, Kobayashi Y, Minamino A, Takenaka S, Nakamura N, Ji XC, Purba ER, Hall M, Yu LJ, Madigan MT, Mizoguchi A, Iwasaki K, Humbel BM, Kimura Y, Wang-Otomo ZY
Commun Biol (2024) 7 pp. 176-176 [ DOI: doi:10.1038/s42003-024-05863-w Pubmed: 38347078 ]
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Bacteriochlorophyll a (911 Da, Ligand)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Reaction center protein l chain (31 kDa, Protein from Allochromatium vinosum DSM 180)
- Spirilloxanthin (596 Da, Ligand)
- Photosynthetic lh1-rc complex from the purple sulfur phototrophic bacterium allochromatium vinosum (Complex from Allochromatium vinosum DSM 180)
- (2s)-3-hydroxypropane-1,2-diyl dihexadecanoate (568 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- (1r)-2-{[{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl (11e)-octadec-11-enoate (749 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Reaction center protein m chain (36 kDa, Protein from Allochromatium vinosum DSM 180)
- Magnesium ion (24 Da, Ligand)
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Photosynthetic reaction center h subunit (28 kDa, Protein from Allochromatium vinosum DSM 180)
- Photosynthetic reaction center cytochrome c subunit (41 kDa, Protein from Allochromatium vinosum DSM 180)
- Ubiquinone-8 (727 Da, Ligand)
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Calcium ion (40 Da, Ligand)
- Menaquinone 8 (717 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Lauryl dimethylamine-n-oxide (229 Da, Ligand)
- Cardiolipin (1 kDa, Ligand)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
Resolution: 2.24 Å
EM Method: Single-particle
Fitted PDBs: 8wdv
Q-score: 0.658
Tani K, Kanno R, Harada A, Kobayashi Y, Minamino A, Takenaka S, Nakamura N, Ji XC, Purba ER, Hall M, Yu LJ, Madigan MT, Mizoguchi A, Iwasaki K, Humbel BM, Kimura Y, Wang-Otomo ZY
Commun Biol (2024) 7 pp. 176-176 [ DOI: doi:10.1038/s42003-024-05863-w Pubmed: 38347078 ]
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Bacteriochlorophyll a (911 Da, Ligand)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Reaction center protein l chain (31 kDa, Protein from Allochromatium vinosum DSM 180)
- Spirilloxanthin (596 Da, Ligand)
- Photosynthetic lh1-rc complex from the purple sulfur phototrophic bacterium allochromatium vinosum (Complex from Allochromatium vinosum DSM 180)
- (2s)-3-hydroxypropane-1,2-diyl dihexadecanoate (568 Da, Ligand)
- Bacteriopheophytin a (889 Da, Ligand)
- (1r)-2-{[{[(2s)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl (11e)-octadec-11-enoate (749 Da, Ligand)
- Fe (iii) ion (55 Da, Ligand)
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Reaction center protein m chain (36 kDa, Protein from Allochromatium vinosum DSM 180)
- Magnesium ion (24 Da, Ligand)
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Photosynthetic reaction center h subunit (28 kDa, Protein from Allochromatium vinosum DSM 180)
- Photosynthetic reaction center cytochrome c subunit (41 kDa, Protein from Allochromatium vinosum DSM 180)
- Ubiquinone-8 (727 Da, Ligand)
- Antenna complex alpha/beta subunit (7 kDa, Protein from Allochromatium vinosum DSM 180)
- Calcium ion (40 Da, Ligand)
- Menaquinone 8 (717 Da, Ligand)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Antenna complex alpha/beta subunit (5 kDa, Protein from Allochromatium vinosum DSM 180)
- Lauryl dimethylamine-n-oxide (229 Da, Ligand)
- Cardiolipin (1 kDa, Ligand)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 6klo
Q-score: 0.586
Yamada T, Yoshida T, Kawamoto A, Mitsuoka K, Iwasaki K, Tsuge H
Nat Struct Mol Biol (2020) 27 pp. 288-296 [ Pubmed: 32123390 DOI: doi:10.1038/s41594-020-0388-6 ]
- Complex structure of iota toxin enzymatic component (ia) and binding component (ib) pore with short stem (570 kDa, Complex from Clostridium perfringens)
- Calcium ion (40 Da, Ligand)
- Iota toxin component ib (74 kDa, Protein from Clostridium perfringens)
- Iota toxin component ia (48 kDa, Protein from Clostridium perfringens)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6klw
Q-score: 0.572
Yamada T, Yoshida T, Kawamoto A, Mitsuoka K, Iwasaki K, Tsuge H
Nat Struct Mol Biol (2020) 27 pp. 288-296 [ Pubmed: 32123390 DOI: doi:10.1038/s41594-020-0388-6 ]
- Complex structure of iota toxin enzymatic component (ia) and binding component (ib) pore with long stem (570 kDa, Complex from Clostridium perfringens)
- Calcium ion (40 Da, Ligand)
- Iota toxin component ib (74 kDa, Protein from Clostridium perfringens)
- Iota toxin component ia (48 kDa, Protein from Clostridium perfringens)
Pore structure of Iota toxin binding component (Ib)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6klx
Q-score: 0.596
Yamada T, Yoshida T, Kawamoto A, Mitsuoka K, Iwasaki K, Tsuge H
Nat Struct Mol Biol (2020) 27 pp. 288-296 [ Pubmed: 32123390 DOI: doi:10.1038/s41594-020-0388-6 ]
- Iota toxin component ib (74 kDa, Protein from Clostridium perfringens)
- Calcium ion (40 Da, Ligand)
- Pore structure of iota toxin binding component (ib) (520 kDa, Complex from Clostridium perfringens)
Cryo-EM structure of T=3 Penaeus vannamei nodavirus
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 6ab6
Q-score: 0.505
Chen NC, Yoshimura M, Miyazaki N, Guan HH, Chuankhayan P, Lin CC, Chen SK, Lin PJ, Huang YC, Iwasaki K, Nakagawa A, Chan SI, Chen CJ
Commun Biol (2019) 2 pp. 72-72 [ DOI: doi:10.1038/s42003-019-0311-z Pubmed: 30820467 ]
- Calcium ion (40 Da, Ligand)
- Capsid protein (40 kDa, Protein from Penaeus vannamei nodavirus)
- Penaeus vannamei nodavirus (Virus from Penaeus vannamei nodavirus)
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6rvv
Q-score: 0.445
Malay AD, Miyazaki N, Biela A, Chakraborti S, Majsterkiewicz K, Stupka I, Kaplan CS, Kowalczyk A, Piette BMAG, Hochberg GKA, Wu D, Wrobel TP, Fineberg A, Kushwah MS, Kelemen M, Vavpetic P, Pelicon P, Kukura P, Benesch JLP, Iwasaki K, Heddle JG
Nature (2019) 569 pp. 438-442 [ Pubmed: 31068697 DOI: doi:10.1038/s41586-019-1185-4 ]
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6rvw
Q-score: 0.449
Malay AD, Miyazaki N, Biela A, Chakraborti S, Majsterkiewicz K, Stupka I, Kaplan CS, Kowalczyk A, Piette BMAG, Hochberg GKA, Wu D, Wrobel TP, Fineberg A, Kushwah MS, Kelemen M, Vavpetic P, Pelicon P, Kukura P, Benesch JLP, Iwasaki K, Heddle JG
Nature (2019) 569 pp. 438-442 [ Pubmed: 31068697 DOI: doi:10.1038/s41586-019-1185-4 ]
Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1
Resolution: 21.0 Å
EM Method: Single-particle
Fitted PDBs: 5gje
Q-score: 0.027
Matoba K, Mihara E, Tamura-Kawakami K, Miyazaki N, Maeda S, Hirai H, Thompson S, Iwasaki K, Takagi J
Cell Rep (2017) 18 pp. 32-40 [ DOI: doi:10.1016/j.celrep.2016.12.017 Pubmed: 28052259 ]
- Phosphate ion (94 Da, Ligand)
- Lrp6 ectodomain (Complex)
- Glycerol (92 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Dickkopf-related protein 1 (9 kDa, Protein from Homo sapiens)
- Low-density lipoprotein receptor-related protein 6 (69 kDa, Protein from Homo sapiens)
- Human dickkopf-related protein1 (Complex)
- Low-density lipoprotein receptor-related protein 6 (69 kDa, Protein from Homo sapiens)
- Human lrp6 ectodomain complexed with full length of dkk1 (210 kDa, Complex from Homo sapiens)
- Water (18 Da, Ligand)