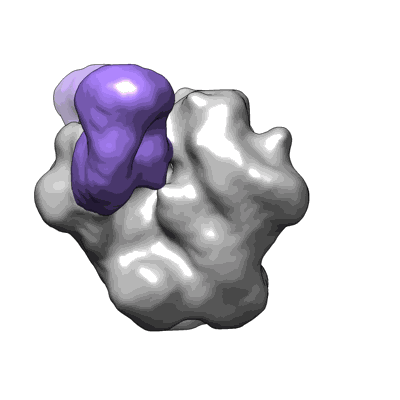
HIV Envelope trimer BG505 SOSIP.664 in complex with wild type CH103 antibody
Liu Q, Parsons R, Wiehe K, Edwards RJ, Saunders KO, Zhang P, Miao H, Williams W, Huang X, Janowska K, Mansouri K, Acharya P, Haynes BF, Lusso P
To Be Published
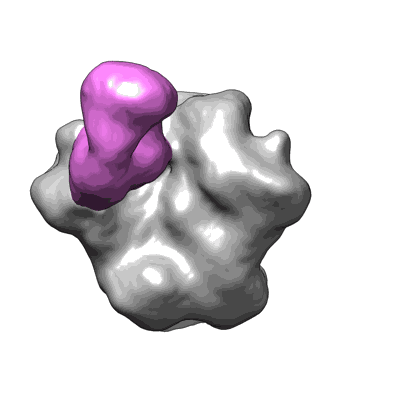
HIV Envelope trimer CH505 SOSIP.664 in complex with wild type CH103 antibody
Liu Q, Parsons R, Wiehe K, Edwards RJ, Saunders KO, Zhang P, Miao H, Williams W, Huang X, Janowska K, Mansouri K, Acharya P, Haynes BF, Lusso P
To Be Published
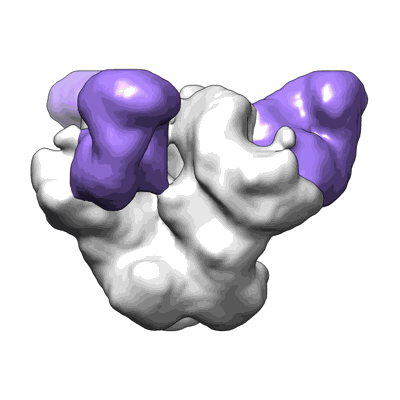
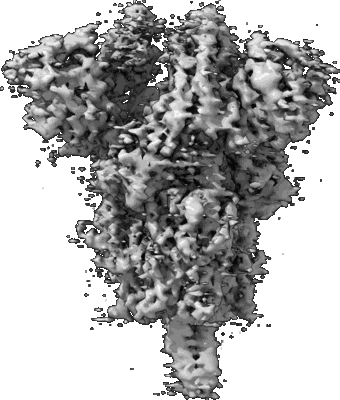
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Resolution: 3.43 Å
EM Method: Single-particle
Fitted PDBs: 7sg4
Q-score: 0.355
Martinez DR, Schafer A, Gobeil S, Li D, De la Cruz G, Parks R, Lu X, Barr M, Stalls V, Janowska K, Beaudoin E, Manne K, Mansouri K, Edwards RJ, Cronin K, Yount B, Anasti K, Montgomery SA, Tang J, Golding H, Shen S, Zhou T, Kwong PD, Graham BS, Mascola JR, Montefiori DC, Alam SM, Sempowski G, Sempowski GD, Khurana S, Wiehe K, Saunders KO, Acharya P, Haynes BF, Baric RS
Sci Transl Med (2022) 14 pp. eabj7125-eabj7125 [ DOI: doi:10.1126/scitranslmed.abj7125 Pubmed: 34726473 ]
- Dh1047 heavy chain (24 kDa, Protein from Homo sapiens)
- Sars-cov s protein in complex with receptor binding domain antibody dh1047 (Complex from Severe acute respiratory syndrome-related coronavirus)
- Dh1047 light chain (24 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus)
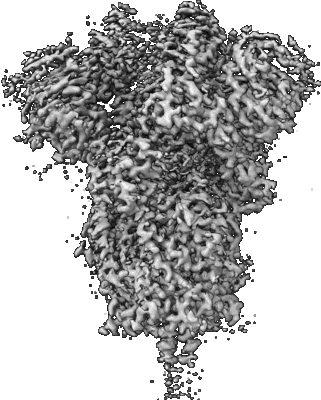
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 7lwi
Q-score: 0.454
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
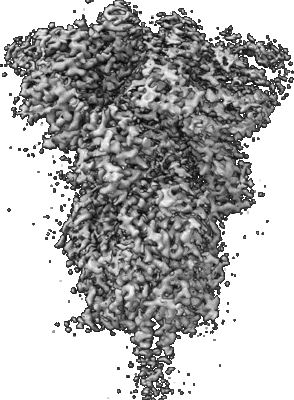
Resolution: 3.24 Å
EM Method: Single-particle
Fitted PDBs: 7lwj
Q-score: 0.363
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
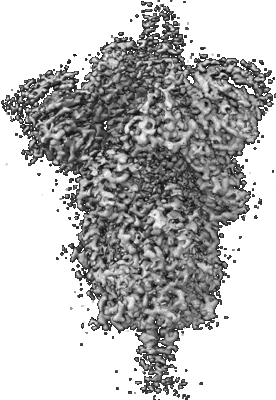
Resolution: 2.92 Å
EM Method: Single-particle
Fitted PDBs: 7lwk
Q-score: 0.501
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
Resolution: 2.84 Å
EM Method: Single-particle
Fitted PDBs: 7lwl
Q-score: 0.515
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
Mink Cluster 5-associated SARS-CoV-2 spike protein (S-GSAS-D614G-delFV) in the 1-RBD up conformation
Resolution: 2.83 Å
EM Method: Single-particle
Fitted PDBs: 7lwm
Q-score: 0.521
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]