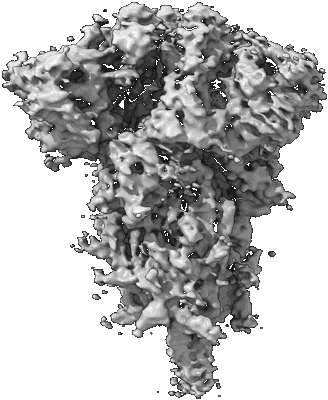
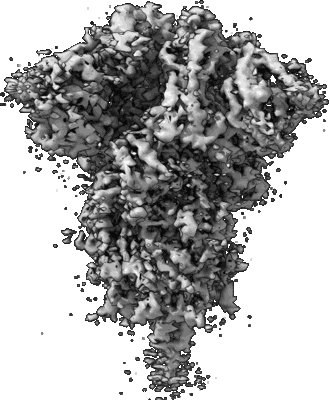
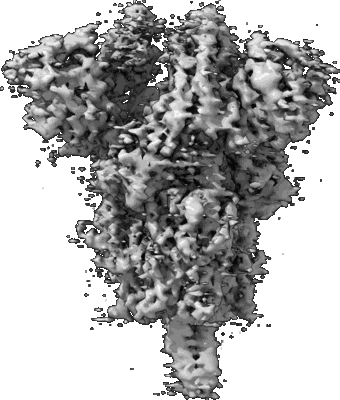
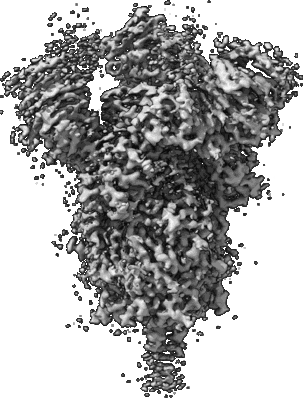
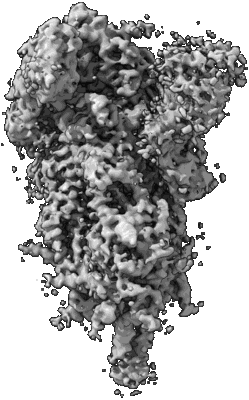
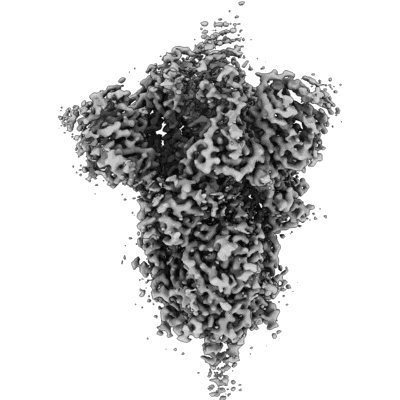Resolution: 3.84 Å
EM Method: Single-particle
Fitted PDBs: 8csa
Q-score: 0.33
Gobeil SM-C, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders K, Edwards RJ, Korber B, Haynes BF, Henderson RC, Acharya P
Science (2021) [ DOI: doi:10.1126/science.abi6226 ]
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 7lwi
Q-score: 0.454
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
Resolution: 3.24 Å
EM Method: Single-particle
Fitted PDBs: 7lwj
Q-score: 0.363
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
Mink Cluster 5-associated SARS-CoV-2 spike protein (S-GSAS-D614G-delFV) in the 2-RBD up conformation
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 7lwp
Q-score: 0.442
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
Resolution: 3.44 Å
EM Method: Single-particle
Fitted PDBs: 7lwq
Q-score: 0.409
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
UK (B.1.1.7) SARS-CoV-2 S-GSAS-D614G variant spike protein in the 3-RBD-down conformation
Resolution: 3.22 Å
EM Method: Single-particle
Fitted PDBs: 7lws
Q-score: 0.473
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Saunders K, Edwards RJ, Haynes BF, Henderson RC, Acharya P
bioRxiv (2021) [ Pubmed: 33758838 DOI: doi:10.1101/2021.03.11.435037 ]
UK (B.1.1.7) SARS-CoV-2 spike protein variant (S-GSAS-B.1.1.7) in the 1-RBD-up conformation
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 7lwt
Q-score: 0.481
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
UK (B.1.1.7) SARS-CoV-2 spike protein variant (S-GSAS-B.1.1.7) in the 1-RBD-up conformation
Resolution: 3.22 Å
EM Method: Single-particle
Fitted PDBs: 7lwu
Q-score: 0.454
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
UK (B.1.1.7) SARS-CoV-2 spike protein variant (S-GSAS-B.1.1.7) in the 1-RBD-up conformation
Resolution: 3.12 Å
EM Method: Single-particle
Fitted PDBs: 7lwv
Q-score: 0.497
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7lww
Q-score: 0.494
Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P
Science (2021) 373 [ DOI: doi:10.1126/science.abi6226 Pubmed: 34168071 ]
- Spike protein trimer; triple mutant (k417n-e484k-n501y) sars-cov-2 spike protein in the 1-rbd-up conformation (s-gsas-d614g-k417n-e484k-n501y) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)