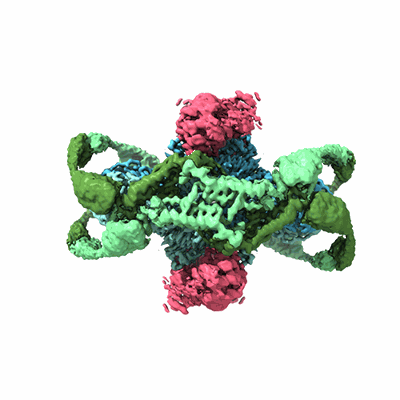
Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex
Resolution: 2.57 Å
EM Method: Single-particle
Fitted PDBs: 8u7i
Q-score: 0.491
Antine SP, Johnson AG, Mooney SE, Leavitt A, Mayer ML, Yirmiya E, Amitai G, Sorek R, Kranzusch PJ
Nature (2024) 625 pp. 360-365 [ Pubmed: 37992757 DOI: doi:10.1038/s41586-023-06855-2 ]
- Gabija protein gajb (57 kDa, Protein from Bacillus cereus VD045)
- Gabija anti-defense 1 (34 kDa, Protein from Bacillus phage phi3T)
- Gad1 octamer (Complex from Bacillus phage phi3T)
- Phage phi3t gad1 bound to the bacillus cereus gabija gajab complex (280 kDa, Complex from Bacillus phage phi3T)
- Endonuclease gaja (78 kDa, Protein from Bacillus cereus VD045)
- Gabija gajab complex (Complex from Bacillus cereus VD045)
Structure of RdrA from Escherichia coli RADAR defense system
Resolution: 2.52 Å
EM Method: Single-particle
Fitted PDBs: 8fnt
Q-score: 0.486
Duncan-Lowey B, Tal N, Johnson AG, Rawson S, Mayer ML, Doron S, Millman A, Melamed S, Fedorenko T, Kacen A, Brandis A, Mehlman T, Amitai G, Sorek R, Kranzusch PJ
Cell (2023) 186 pp. 987- [ DOI: doi:10.1016/j.cell.2023.01.012 Pubmed: 36764290 ]
- E. coli rdra complex (Complex from Escherichia coli)
- Archaeal atpase (107 kDa, Protein from Escherichia coli)
Map of RdrA from Escherichia coli RADAR defense system in single-split conformation
Duncan-Lowey B, Tal N, Johnson AG, Rawson S, Mayer ML, Doron S, Millman A, Melamed S, Fedorenko T, Kacen A, Brandis A, Mehlman T, Amitai G, Sorek R, Kranzusch PJ
Cell (2023) 186 pp. 987-998.e15 [ DOI: doi:10.1016/j.cell.2023.01.012 Pubmed: 36764290 ]
Map of RdrA from Escherichia coli RADAR defense system in double-split conformation
Duncan-Lowey B, Tal N, Johnson AG, Rawson S, Mayer ML, Doron S, Millman A, Melamed S, Fedorenko T, Kacen A, Brandis A, Mehlman T, Amitai G, Sorek R, Kranzusch PJ
Cell (2023) 186 pp. 987-998.e15 [ DOI: doi:10.1016/j.cell.2023.01.012 Pubmed: 36764290 ]
Structure of RdrA from Streptococcus suis RADAR defense system
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8fnu
Q-score: 0.572
Duncan-Lowey B, Tal N, Johnson AG, Rawson S, Mayer ML, Doron S, Millman A, Melamed S, Fedorenko T, Kacen A, Brandis A, Mehlman T, Amitai G, Sorek R, Kranzusch PJ
Cell (2023) 186 pp. 987- [ DOI: doi:10.1016/j.cell.2023.01.012 Pubmed: 36764290 ]
- Rdra heptameric complex (Complex from Streptococcus suis)
- Kap ntpase domain-containing protein (106 kDa, Protein from Streptococcus suis)
Structure of RdrB from Escherichia coli RADAR defense system
Resolution: 2.11 Å
EM Method: Single-particle
Fitted PDBs: 8fnv
Q-score: 0.664
Duncan-Lowey B, Tal N, Johnson AG, Rawson S, Mayer ML, Doron S, Millman A, Melamed S, Fedorenko T, Kacen A, Brandis A, Mehlman T, Amitai G, Sorek R, Kranzusch PJ
Cell (2023) 186 pp. 987- [ DOI: doi:10.1016/j.cell.2023.01.012 Pubmed: 36764290 ]
- Escherichia coli rdrb dodecamer (Complex from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Adenosine deaminase (92 kDa, Protein from Escherichia coli)