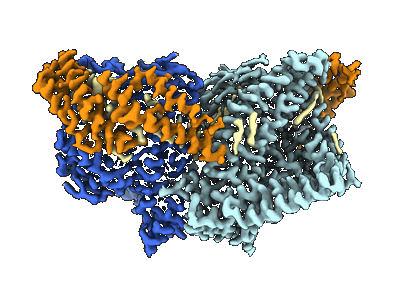
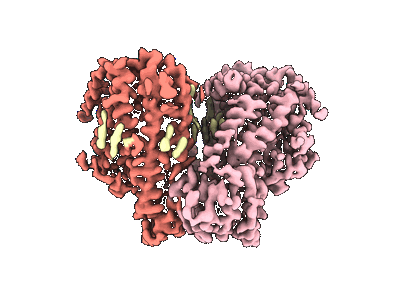
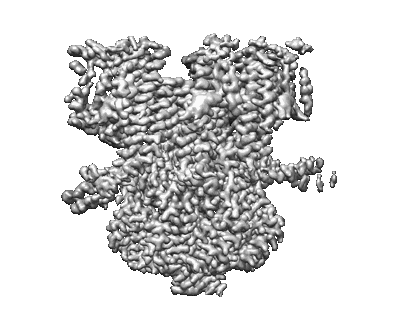
Mechanically activated ion channel OSCA3.1 in nanodiscs
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8u53
Q-score: 0.54
Jojoa-Cruz S, Dubin AE, Lee WH, Ward AB
eLife (2024) 12 [ DOI: doi:10.7554/eLife.93147 Pubmed: 38592763 ]
- Palmitic acid (256 Da, Ligand)
- Osca3.1 dimer in nanodisc (164 kDa, Complex from Arabidopsis thaliana)
- Csc1-like protein erd4 (81 kDa, Protein from Arabidopsis thaliana)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- [(2r)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl] octadecanoate (481 Da, Ligand)
Structure of mechanically activated ion channel OSCA1.2 in peptidiscs
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8t56
Q-score: 0.584
Jojoa-Cruz S, Burendei B, Lee WH, Ward AB
Structure (2024) 32 pp. 157- [ Pubmed: 38103547 DOI: doi:10.1016/j.str.2023.11.009 ]
- Palmitic acid (256 Da, Ligand)
- Osca1.2 (Complex from Arabidopsis thaliana)
- Calcium permeable stress-gated cation channel 1 (89 kDa, Protein from Arabidopsis thaliana)
- Osca1.2 in peptidisc (176 kDa, Complex)
- Nspr peptide (4 kDa, Protein from synthetic construct)
- Peptidisc peptide (Complex from synthetic construct)
- Cholesterol (386 Da, Ligand)
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (760 Da, Ligand)
- Cholesterol hemisuccinate (486 Da, Ligand)
Structure of mechanically activated ion channel OSCA2.3 in peptidiscs
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8t57
Q-score: 0.53
Jojoa-Cruz S, Burendei B, Lee WH, Ward AB
Structure (2024) 32 pp. 157- [ Pubmed: 38103547 DOI: doi:10.1016/j.str.2023.11.009 ]
- Palmitic acid (256 Da, Ligand)
- Cholesterol (386 Da, Ligand)
- Csc1-like protein hyp1 (80 kDa, Protein from Arabidopsis thaliana)
- Osca2.3 in peptidisc (159 kDa, Complex from Arabidopsis thaliana)
Cryo-EM of Mechanosensitive Ion Channel Flycatcher1 in GDN
Jojoa-Cruz S, Saotome K, Tsui CCA, Lee WH, Sansom MSP, Murthy SE, Patapoutian A, Ward AB
Nat Commun (2022) 13 [ Pubmed: 35165281 DOI: 10.1038/s41467-022-28511-5 ]
Image sets:
Composite Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7n5d
Q-score: 0.489
Jojoa-Cruz S, Saotome K, Tsui CCA, Lee WH, Sansom MSP, Murthy SE, Patapoutian A, Ward AB
Nat Commun (2022) 13 pp. 850-850 [ Pubmed: 35165281 DOI: doi:10.1038/s41467-022-28511-5 ]
- Palmitic acid (256 Da, Ligand)
- Flyc1 (607 kDa, Cellular component from Dionaea muscipula)
- Mechanosensitive ion channel flycatcher1 (86 kDa, Protein from Dionaea muscipula)
Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 7n5e
Q-score: 0.453
Jojoa-Cruz S, Saotome K, Tsui CCA, Lee WH, Sansom MSP, Murthy SE, Patapoutian A, Ward AB
Nat Commun (2022) 13 pp. 850-850 [ Pubmed: 35165281 DOI: doi:10.1038/s41467-022-28511-5 ]
- Palmitic acid (256 Da, Ligand)
- Flyc1 (607 kDa, Cellular component from Dionaea muscipula)
- Mechanosensitive ion channel flycatcher1 (86 kDa, Protein from Dionaea muscipula)
Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Down' conformation in GDN
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7n5f
Q-score: 0.44
Jojoa-Cruz S, Saotome K, Tsui CCA, Lee WH, Sansom MSP, Murthy SE, Patapoutian A, Ward AB
Nat Commun (2022) 13 pp. 850-850 [ Pubmed: 35165281 DOI: doi:10.1038/s41467-022-28511-5 ]
- Palmitic acid (256 Da, Ligand)
- Flyc1 (607 kDa, Cellular component from Dionaea muscipula)
- Mechanosensitive ion channel flycatcher1 (86 kDa, Protein from Dionaea muscipula)
Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Up' conformation in GDN
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 7n5g
Q-score: 0.496
Jojoa-Cruz S, Saotome K, Tsui CCA, Lee WH, Sansom MSP, Murthy SE, Patapoutian A, Ward AB
Nat Commun (2022) 13 pp. 850-850 [ Pubmed: 35165281 DOI: doi:10.1038/s41467-022-28511-5 ]
- Palmitic acid (256 Da, Ligand)
- Flyc1 (607 kDa, Cellular component from Dionaea muscipula)
- Mechanosensitive ion channel flycatcher1 (86 kDa, Protein from Dionaea muscipula)
Cryo-EM map of mechanically activated ion channel OSCA1.2 in nanodisc
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 6mgv
Q-score: 0.502
Jojoa Cruz S, Saotome K, Murthy SE, Tsui CCA, Sansom MS, Patapoutian A, Ward AB
eLife (2018) 7 [ DOI: doi:10.7554/eLife.41845 Pubmed: 30382939 ]
- Calcium permeable stress-gated cation channel 1 (89 kDa, Protein from Arabidopsis thaliana)
- Osca1.2 (88 kDa, Cellular component from Arabidopsis thaliana)
Cryo-EM map of mechanically activated ion channel OSCA1.2 in LMNG
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 6mgw
Q-score: 0.41
Jojoa Cruz S, Saotome K, Murthy SE, Tsui CCA, Sansom MS, Patapoutian A, Ward AB
eLife (2018) 7 [ DOI: doi:10.7554/eLife.41845 Pubmed: 30382939 ]
- Calcium permeable stress-gated cation channel 1 (89 kDa, Protein from Arabidopsis thaliana)
- Osca1.2 (88 kDa, Cellular component from Arabidopsis thaliana)