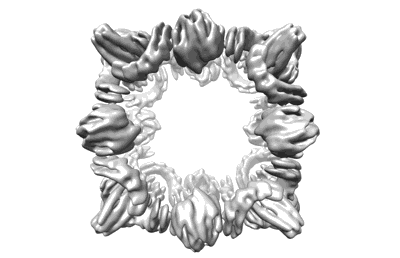
Backbone model of de novo-designed chlorophyll-binding nanocage O32-15
Resolution: 6.5 Å
EM Method: Single-particle
Fitted PDBs: 8glt
Q-score: 0.213
Ennist NM, Wang S, Kennedy MA, Curti M, Sutherland GA, Vasilev C, Redler RL, Maffeis V, Shareef S, Sica AV, Hua AS, Deshmukh AP, Moyer AP, Hicks DR, Swartz AZ, Cacho RA, Novy N, Bera AK, Kang A, Sankaran B, Johnson MP, Phadkule A, Reppert M, Ekiert D, Bhabha G, Stewart L, Caram JR, Stoddard BL, Romero E, Hunter CN, Baker D
Nat Chem Biol (2024) 20 pp. 906-915 [ Pubmed: 38831036 DOI: doi:10.1038/s41589-024-01626-0 ]
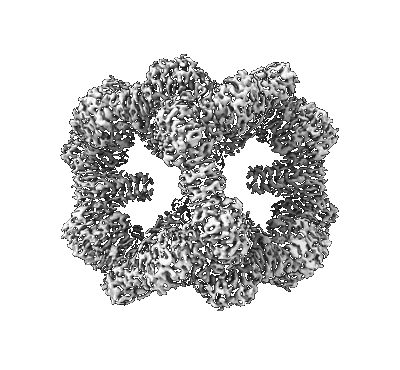
Chlorophyll-binding region of de novo-designed nanocage O32-15
Ennist NM, Wang S, Kennedy MA, Curti M, Sutherland GA, Vasilev C, Redler RL, Maffeis V, Shareef S, Sica AV, Hua AS, Deshmukh AP, Moyer AP, Hicks DR, Swartz AZ, Cacho RA, Novy N, Bera AK, Kang A, Sankaran B, Johnson MP, Phadkule A, Reppert M, Ekiert D, Bhabha G, Stewart L, Caram JR, Stoddard BL, Romero E, Hunter CN, Baker D
Nat Chem Biol (2024) 20 pp. 906-915 [ Pubmed: 38831036 DOI: doi:10.1038/s41589-024-01626-0 ]
Twistless helix 12 repeat ring design R12B
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
- R12b (Protein from synthetic construct)
- Twistless helix 12 repeat ring design r12b (155 kDa, Complex from synthetic construct)
Cryo-EM structure of synthetic tetrameric building block sC4
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 8gel
Q-score: 0.439
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
- Sc4 (57 kDa, Protein from synthetic construct)
- De novo designed tetramer sc4 (Complex from synthetic construct)
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Resolution: 4.05 Å
EM Method: Single-particle
Fitted PDBs: 8tl7
Q-score: 0.338
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
- Computationally designed protein (78 kDa, Protein from synthetic construct)
- T3 tetrahedral cage (Complex from unidentified)
Computational Designed Nanocage O43_129
Resolution: 6.77 Å
EM Method: Single-particle
Fitted PDBs: 8v2d
Q-score: 0.201
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
- O43_129 component a (33 kDa, Protein from synthetic construct)
- Nanocage o43_129 (1 MDa, Complex from unidentified)
- O43_129 component b (34 kDa, Protein from synthetic construct)
Computational Designed Nanocage O43_129_+4
Resolution: 6.4 Å
EM Method: Single-particle
Fitted PDBs: 8v3b
Q-score: 0.217
Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D
Nature (2024) 627 pp. 898-904 [ DOI: doi:10.1038/s41586-024-07188-4 Pubmed: 38480887 ]
- O43_129_+4 component b (46 kDa, Protein from synthetic construct)
- O43_129_+4 component a (33 kDa, Protein from synthetic construct)
- Computational designed nanocage o43_129_+4 (Complex from unidentified)
Accurate computational design of genetically encoded 3D protein crystals
Resolution: 3.34 Å
EM Method: Single-particle
Fitted PDBs: 8cwy
Q-score: 0.452
Li Z, Wang S, Nattermann U, Bera AK, Borst AJ, Yaman MY, Bick MJ, Yang EC, Sheffler W, Lee B, Seifert S, Hura GL, Nguyen H, Kang A, Dalal R, Lubner JM, Hsia Y, Haddox H, Courbet A, Dowling Q, Miranda M, Favor A, Etemadi A, Edman NI, Yang W, Weidle C, Sankaran B, Negahdari B, Ross MB, Ginger DS, Baker D
Nat Mater (2023) 22 pp. 1556-1563 [ Pubmed: 37845322 DOI: doi:10.1038/s41563-023-01683-1 ]
- T32-15-1 (49 kDa, Protein from synthetic construct)
- T32-15 (Complex from Escherichia coli)
- T32-15-2 (9 kDa, Protein from synthetic construct)
CryoEM Structure of Computationally Designed Nanocage O32-ZL4
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8szz
Q-score: 0.591
Li Z, Wang S, Nattermann U, Bera AK, Borst AJ, Yaman MY, Bick MJ, Yang EC, Sheffler W, Lee B, Seifert S, Hura GL, Nguyen H, Kang A, Dalal R, Lubner JM, Hsia Y, Haddox H, Courbet A, Dowling Q, Miranda M, Favor A, Etemadi A, Edman NI, Yang W, Weidle C, Sankaran B, Negahdari B, Ross MB, Ginger DS, Baker D
Nat Mater (2023) 22 pp. 1556-1563 [ Pubmed: 37845322 DOI: doi:10.1038/s41563-023-01683-1 ]
- O32-zl4 component a (8 kDa, Protein from synthetic construct)
- O32-zl4 (764 kDa, Complex from Thermotoga maritima)
- Sodium ion (22 Da, Ligand)
- O32-zl4 component b (23 kDa, Protein from Thermotoga maritima)
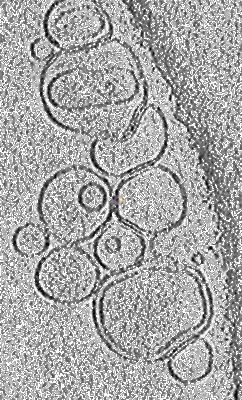
Representative cryo-electron tomogram of pRLB-540 mixed with liposomes at pH 8.0
Goldbach N, Benna I, Wicky BIM, Croft JT, Carter L, Bera AK, Nguyen H, Kang A, Sankaran B, Yang EC, Lee KK, Baker D
Protein Sci (2023) 32 pp. e4769-e4769 [ DOI: doi:10.1002/pro.4769 Pubmed: 37632837 ]