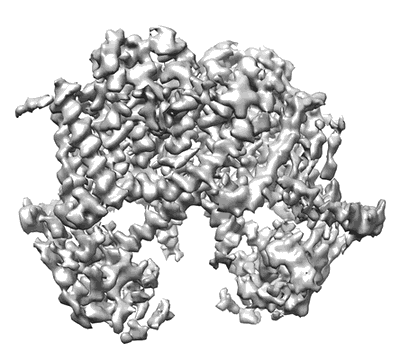
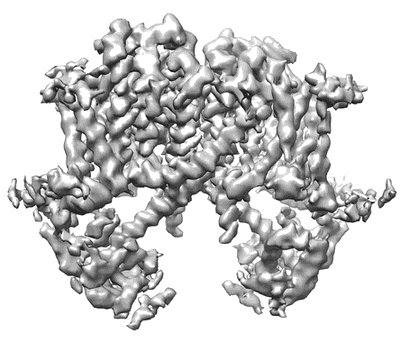
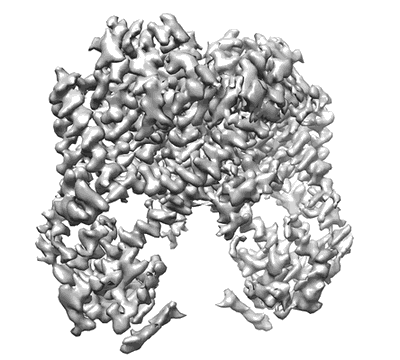Cryo-EM structure of the HIV-1 restriction factor human SERINC3
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 7ru6
Q-score: 0.323
Leonhardt SA, Purdy MD, Grover JR, Yang Z, Poulos S, McIntire WE, Tatham EA, Erramilli SK, Nosol K, Lai KK, Ding S, Lu M, Uchil PD, Finzi A, Rein A, Kossiakoff AA, Mothes W, Yeager M
Nat Commun (2023) 14 pp. 4368-4368 [ DOI: doi:10.1038/s41467-023-39262-2 Pubmed: 37474505 ]
- Sia (25 kDa, Protein from synthetic construct)
- Hserinc3-sia (55 kDa, Complex from Homo sapiens)
- Serine incorporator 3 (55 kDa, Protein from Homo sapiens)
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 7rug
Q-score: 0.244
Leonhardt SA, Purdy MD, Grover JR, Yang Z, Poulos S, McIntire WE, Tatham EA, Erramilli SK, Nosol K, Lai KK, Ding S, Lu M, Uchil PD, Finzi A, Rein A, Kossiakoff AA, Mothes W, Yeager M
Nat Commun (2023) 14 pp. 4368-4368 [ DOI: doi:10.1038/s41467-023-39262-2 Pubmed: 37474505 ]
- Hserinc3-deltaicl4-sia (52 kDa, Complex from Homo sapiens)
- Sia (25 kDa, Protein from synthetic construct)
- Serine incorporator 3 (52 kDa, Protein from Homo sapiens)
Structure of AtTPC1(DDE) reconstituted in saposin A with cat06 Fab
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 6e1k
Q-score: 0.408
Kintzer AF, Green EM, Dominik PK, Bridges M, Armache JP, Deneka D, Kim SS, Hubbell W, Kossiakoff AA, Cheng Y, Stroud RM
PNAS (2018) 115 pp. E9095-E9104 [ DOI: doi:10.1073/pnas.1805651115 Pubmed: 30190435 ]
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Two pore calcium channel protein 1 (84 kDa, Protein from Arabidopsis thaliana)
- Cat06 light chain (23 kDa, Protein from Homo sapiens)
- Cat06 heavy chain (24 kDa, Protein from Homo sapiens)
- Attpc1(dde) reconstituted in saposin a (Complex from Arabidopsis thaliana)
- Calcium ion (40 Da, Ligand)
Structure of AtTPC1(DDE) reconstituted in saposin A
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 6e1m
Q-score: 0.462
Kintzer AF, Green EM, Dominik PK, Bridges M, Armache JP, Deneka D, Kim SS, Hubbell W, Kossiakoff AA, Cheng Y, Stroud RM
PNAS (2018) 115 pp. E9095-E9104 [ DOI: doi:10.1073/pnas.1805651115 Pubmed: 30190435 ]
- 1,2-diacyl-glycerol-3-sn-phosphate (704 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Attpc1(dde) (Complex from Arabidopsis thaliana)
- Two pore calcium channel protein 1 (84 kDa, Protein from Arabidopsis thaliana)
- Water (18 Da, Ligand)
- Calcium ion (40 Da, Ligand)
Structure of AtTPC1(DDE) in state 1
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6e1n
Q-score: 0.398
Kintzer AF, Green EM, Dominik PK, Bridges M, Armache JP, Deneka D, Kim SS, Hubbell W, Kossiakoff AA, Cheng Y, Stroud RM
PNAS (2018) 115 pp. E9095-E9104 [ DOI: doi:10.1073/pnas.1805651115 Pubmed: 30190435 ]
- 1,2-diacyl-glycerol-3-sn-phosphate (704 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Attpc1(dde) (Complex from Arabidopsis thaliana)
- Two pore calcium channel protein 1 (84 kDa, Protein from Arabidopsis thaliana)
- Water (18 Da, Ligand)
- Calcium ion (40 Da, Ligand)
Structure of AtTPC1(DDE) in state 2
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6e1p
Q-score: 0.355
Kintzer AF, Green EM, Dominik PK, Bridges M, Armache JP, Deneka D, Kim SS, Hubbell W, Kossiakoff AA, Cheng Y, Stroud RM
PNAS (2018) 115 pp. E9095-E9104 [ DOI: doi:10.1073/pnas.1805651115 Pubmed: 30190435 ]
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Attpc1(dde) (Complex from Arabidopsis thaliana)
- Two pore calcium channel protein 1 (84 kDa, Protein from Arabidopsis thaliana)
- Calcium ion (40 Da, Ligand)
Cryo-EM structure of human insulin degrading enzyme in complex with insulin
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 6bf8
Q-score: 0.153
Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Wang A, Farcasanu M, Woods VA, McCord LA, Lee D, Shang W, Deprez-Poulain R, Deprez B, Liu DR, Koide A, Koide S, Kossiakoff AA, Li S, Carragher B, Potter CS, Tang WJ
eLife (2018) 7 [ DOI: doi:10.7554/eLife.33572 Pubmed: 29596046 ]
- Insulin-degrading enzyme (111 kDa, Protein from Homo sapiens)
- Insulin degrading enzyme (100 kDa, Complex from Homo sapiens)
Cryo-EM structure of human insulin degrading enzyme
Resolution: 6.5 Å
EM Method: Single-particle
Fitted PDBs: 6bf6
Q-score: 0.162
Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Wang A, Farcasanu M, Woods VA, McCord LA, Lee D, Shang W, Deprez-Poulain R, Deprez B, Liu DR, Koide A, Koide S, Kossiakoff AA, Li S, Carragher B, Potter CS, Tang WJ
eLife (2018) 7 [ DOI: doi:10.7554/eLife.33572 Pubmed: 29596046 ]
- Insulin-degrading enzyme (111 kDa, Protein from Homo sapiens)
- Insulin degrading enzyme (100 kDa, Complex from Homo sapiens)
Resolution: 6.5 Å
EM Method: Single-particle
Fitted PDBs: 6bf7
Q-score: 0.16
Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Wang A, Farcasanu M, Woods VA, McCord LA, Lee D, Shang W, Deprez-Poulain R, Deprez B, Liu DR, Koide A, Koide S, Kossiakoff AA, Li S, Carragher B, Potter CS, Tang WJ
eLife (2018) 7 [ DOI: doi:10.7554/eLife.33572 Pubmed: 29596046 ]
- Fab h11-e light chain (23 kDa, Protein from Homo sapiens)
- Insulin-degrading enzyme (111 kDa, Protein from Homo sapiens)
- Insulin degrading enzyme (100 kDa, Complex from Homo sapiens)
- Fab h11-e heavy chain (23 kDa, Protein from Homo sapiens)
Resolution: 7.2 Å
EM Method: Single-particle
Fitted PDBs: 6bf9
Q-score: 0.156
Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Wang A, Farcasanu M, Woods VA, McCord LA, Lee D, Shang W, Deprez-Poulain R, Deprez B, Liu DR, Koide A, Koide S, Kossiakoff AA, Li S, Carragher B, Potter CS, Tang WJ
eLife (2018) 7 [ DOI: doi:10.7554/eLife.33572 Pubmed: 29596046 ]
- Fab h11-e light chain (23 kDa, Protein from Homo sapiens)
- Insulin-degrading enzyme (111 kDa, Protein from Homo sapiens)
- Insulin degrading enzyme (100 kDa, Complex from Homo sapiens)
- Fab h11-e heavy chain (23 kDa, Protein from Homo sapiens)