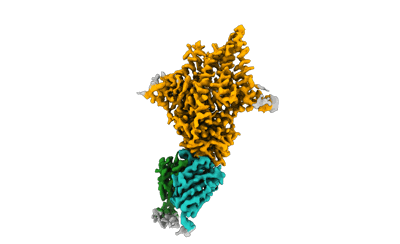
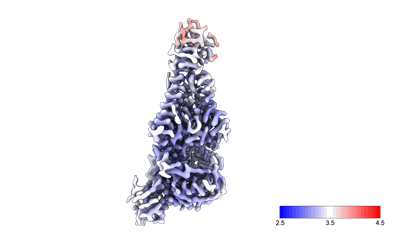
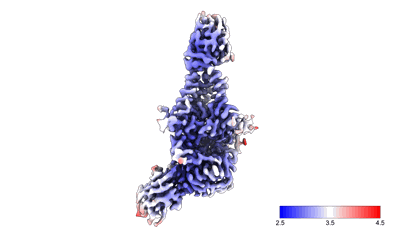
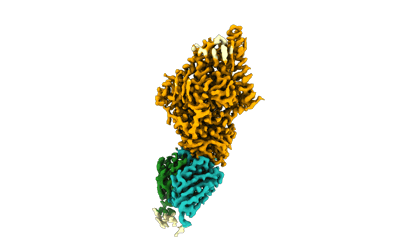Xenopus laevis hyaluronan synthase 1
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8smm
Q-score: 0.504
Gorniak I, Stephens Z, Erramilli SK, Gawda T, Kossiakoff AA, Zimmer J
Nat Struct Mol Biol (2025) 32 pp. 161-171 [ Pubmed: 39322765 DOI: doi:10.1038/s41594-024-01389-1 ]
- Xenopus laevis hyaluronan synthase 1 in complex with a fab molecule (118 kDa, Complex from Xenopus laevis)
- Fab15 heavy chain (24 kDa, Protein from Homo sapiens)
- Hyaluronan synthase 1 (70 kDa, Protein from Xenopus laevis)
- Fab15 light chain (23 kDa, Protein from Homo sapiens)
Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8smn
Q-score: 0.557
Gorniak I, Stephens Z, Erramilli SK, Gawda T, Kossiakoff AA, Zimmer J
Nat Struct Mol Biol (2025) 32 pp. 161-171 [ Pubmed: 39322765 DOI: doi:10.1038/s41594-024-01389-1 ]
- Hyaluronan synthase 1 (70 kDa, Protein from Xenopus laevis)
- Fab15 heavy chain (24 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Xenopus laevis hyaluronan synthase 1 in complex with a fab molecule, nascent ha polymer bound state (118 kDa, Complex from Xenopus laevis)
- Fab15 light chain (23 kDa, Protein from Homo sapiens)
Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8smp
Q-score: 0.511
Gorniak I, Stephens Z, Erramilli SK, Gawda T, Kossiakoff AA, Zimmer J
Nat Struct Mol Biol (2025) 32 pp. 161-171 [ Pubmed: 39322765 DOI: doi:10.1038/s41594-024-01389-1 ]
- Uridine-5'-diphosphate (404 Da, Ligand)
- Hyaluronan synthase 1 (70 kDa, Protein from Xenopus laevis)
- Xenopus laevis hyaluronan synthase 1, udp-bound, gating loop inserted state (118 kDa, Complex from Xenopus laevis)
- Fab15 heavy chain (24 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Fab15 light chain (23 kDa, Protein from Homo sapiens)
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8snc
Q-score: 0.509
Gorniak I, Stephens Z, Erramilli SK, Gawda T, Kossiakoff AA, Zimmer J
Nat Struct Mol Biol (2025) 32 pp. 161-171 [ Pubmed: 39322765 DOI: doi:10.1038/s41594-024-01389-1 ]
- Hyaluronan synthase (65 kDa, Protein from Paramecium bursaria Chlorella virus CZ-2)
- Nanobody 872 (14 kDa, Protein from Lama glama)
- Nanobody 881 (15 kDa, Protein from Lama glama)
- Ternary complex of chlorella virus hyaluronan synthase bound to nanobodies 872 and 886 (95 MDa, Complex from Paramecium bursaria Chlorella virus 1)
- Manganese (ii) ion (54 Da, Ligand)
- Cholesterol hemisuccinate (486 Da, Ligand)
- Water (18 Da, Ligand)
Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8snd
Q-score: 0.512
Gorniak I, Stephens Z, Erramilli SK, Gawda T, Kossiakoff AA, Zimmer J
Nat Struct Mol Biol (2025) 32 pp. 161-171 [ Pubmed: 39322765 DOI: doi:10.1038/s41594-024-01389-1 ]
- Nanobody 872 (14 kDa, Protein from Lama glama)
- Ternary complex of chlorella virus hyaluronan synthase bound to nanobodies 872 and 881 (97 MDa, Complex from Paramecium bursaria Chlorella virus 1)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Uridine-5'-diphosphate-glucuronic acid (580 Da, Ligand)
- Hyaluronan synthase (65 kDa, Protein from Paramecium bursaria Chlorella virus CZ-2)
- Manganese (ii) ion (54 Da, Ligand)
- Cholesterol hemisuccinate (486 Da, Ligand)
- Nanobody 881 (15 kDa, Protein from Lama glama)
- Water (18 Da, Ligand)
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer and UDP
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8sne
Q-score: 0.546
Gorniak I, Stephens Z, Erramilli SK, Gawda T, Kossiakoff AA, Zimmer J
Nat Struct Mol Biol (2025) 32 pp. 161-171 [ Pubmed: 39322765 DOI: doi:10.1038/s41594-024-01389-1 ]
- Hyaluronan synthase (65 kDa, Protein from Paramecium bursaria Chlorella virus CZ-2)
- Uridine-5'-diphosphate (404 Da, Ligand)
- Nanobody 872 (14 kDa, Protein from Lama glama)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Nanobody 886 (14 kDa, Protein from Lama glama)
- Ternary complex of chlorella virus hyaluronan synthase bound to nanobodies 872 and 886 (95 MDa, Complex from Paramecium bursaria Chlorella virus 1)
- Manganese (ii) ion (54 Da, Ligand)
- Cholesterol hemisuccinate (486 Da, Ligand)
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound
Resolution: 2.77 Å
EM Method: Single-particle
Fitted PDBs: 8vzn
Q-score: 0.583
Cater RJ, Mukherjee D, Gil-Iturbe E, Erramilli SK, Chen T, Koo K, Santander N, Reckers A, Kloss B, Gawda T, Choy BC, Zhang Z, Katewa A, Larpthaveesarp A, Huang EJ, Mooney SWJ, Clarke OB, Yee SW, Giacomini KM, Kossiakoff AA, Quick M, Arnold T, Mancia F
Nature (2024) 629 pp. 704-709 [ DOI: doi:10.1038/s41586-024-07326-y Pubmed: 38693257 ]
- Flvcr2 in the inward-facing state complexed with choline and fab flv9 (110 kDa, Complex from Mus musculus)
- Feline leukemia virus subgroup c cellular receptor 2 (60 kDa, Protein from Mus musculus)
- Fab flv9 heavy chain (25 kDa, Protein from synthetic construct)
- Fab flv9 light chain (23 kDa, Protein from synthetic construct)
- Choline ion (104 Da, Ligand)
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound
Resolution: 2.49 Å
EM Method: Single-particle
Fitted PDBs: 8vzo
Q-score: 0.637
Cater RJ, Mukherjee D, Gil-Iturbe E, Erramilli SK, Chen T, Koo K, Santander N, Reckers A, Kloss B, Gawda T, Choy BC, Zhang Z, Katewa A, Larpthaveesarp A, Huang EJ, Mooney SWJ, Clarke OB, Yee SW, Giacomini KM, Kossiakoff AA, Quick M, Arnold T, Mancia F
Nature (2024) 629 pp. 704-709 [ DOI: doi:10.1038/s41586-024-07326-y Pubmed: 38693257 ]
- Flvcr2 in the outward-facing state complexed with choline and fab flv23 (110 kDa, Complex from Mus musculus)
- Feline leukemia virus subgroup c cellular receptor 2 (60 kDa, Protein from Mus musculus)
- Fab flv23 heavy chain (25 kDa, Protein from synthetic construct)
- Fab flv23 light chain (23 kDa, Protein from synthetic construct)
- Choline ion (104 Da, Ligand)
Cryo-EM structure of the HIV-1 restriction factor human SERINC3
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 7ru6
Q-score: 0.323
Leonhardt SA, Purdy MD, Grover JR, Yang Z, Poulos S, McIntire WE, Tatham EA, Erramilli SK, Nosol K, Lai KK, Ding S, Lu M, Uchil PD, Finzi A, Rein A, Kossiakoff AA, Mothes W, Yeager M
Nat Commun (2023) 14 pp. 4368-4368 [ Pubmed: 37474505 DOI: doi:10.1038/s41467-023-39262-2 ]
- Serine incorporator 3 (55 kDa, Protein from Homo sapiens)
- Sia (25 kDa, Protein from synthetic construct)
- Hserinc3-sia (55 kDa, Complex from Homo sapiens)
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 7rug
Q-score: 0.244
Leonhardt SA, Purdy MD, Grover JR, Yang Z, Poulos S, McIntire WE, Tatham EA, Erramilli SK, Nosol K, Lai KK, Ding S, Lu M, Uchil PD, Finzi A, Rein A, Kossiakoff AA, Mothes W, Yeager M
Nat Commun (2023) 14 pp. 4368-4368 [ Pubmed: 37474505 DOI: doi:10.1038/s41467-023-39262-2 ]
- Sia (25 kDa, Protein from synthetic construct)
- Serine incorporator 3 (52 kDa, Protein from Homo sapiens)
- Hserinc3-deltaicl4-sia (52 kDa, Complex from Homo sapiens)