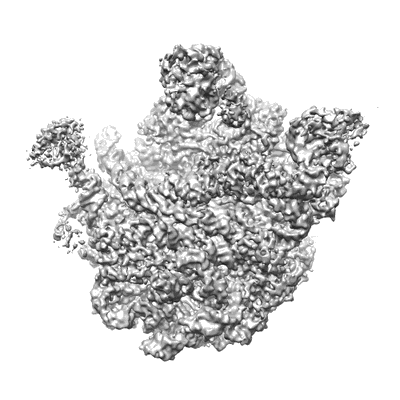
Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7ext
Q-score: 0.11
Zheng L, Zheng Z, Li X, Wang G, Zhang K, Wei P, Zhao J, Gao N
Nat Commun (2021) 12 pp. 5497-5497 [ DOI: doi:10.1038/s41467-021-25813-y Pubmed: 34535665 ]
- Phycobilisome 32.3 kda linker polypeptide, phycocyanin-associated, rod (32 kDa, Protein from Synechococcus sp. PCC 7002)
- Allophycocyanin subunit alpha-b (17 kDa, Protein from Synechococcus sp. PCC 7002)
- C-phycocyanin subunit alpha (17 kDa, Protein from Synechococcus sp. PCC 7002)
- Phycocyanobilin (588 Da, Ligand)
- Phycobilisome 7.8 kda linker polypeptide, allophycocyanin-associated, core (7 kDa, Protein from Synechococcus sp. PCC 7002)
- C-phycocyanin subunit beta (18 kDa, Protein from Synechococcus sp. PCC 7002)
- Phycobiliprotein apce (99 kDa, Protein from Synechococcus sp. PCC 7002)
- Allophycocyanin alpha subunit (17 kDa, Protein from Synechococcus sp. PCC 7002)
- Allophycocyanin beta subunit (17 kDa, Protein from Synechococcus sp. PCC 7002)
- Allophycocyanin subunit beta-18 (18 kDa, Protein from Synechococcus sp. PCC 7002)
- Phycobilisome rod-core linker polypeptide cpcg (28 kDa, Protein from Synechococcus sp. PCC 7002)
- Cryo-em structure of cyanobacterial phycobilisome from synechococcus sp. pcc 7002 (6 MDa, Complex from Synechococcus sp. PCC 7002)
Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7eyd
Q-score: 0.171
Zheng L, Zheng Z, Li X, Wang G, Zhang K, Wei P, Zhao J, Gao N
Nat Commun (2021) 12 pp. 5497-5497 [ DOI: doi:10.1038/s41467-021-25813-y Pubmed: 34535665 ]
- Phycobilisome rod-core linker polypeptide cpcg4 (29 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Allophycocyanin subunit beta (17 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Allophycocyanin subunit alpha-b (17 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Cryo-em structure of cyanobacterial phycobilisome from anabaena sp. pcc 7120 (7 MDa, Complex from Nostoc sp. PCC 7120 = FACHB-418)
- Phycobilisome rod-core linker polypeptide cpcg1 (31 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Phycobilisome 7.8 kda linker polypeptide, allophycocyanin-associated, core (7 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Phycocyanobilin (588 Da, Ligand)
- Phycobilisome 32.1 kda linker polypeptide, phycocyanin-associated, rod (32 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Allophycocyanin subunit beta-18 (18 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- C-phycocyanin beta subunit (18 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Phycobiliprotein apce (127 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Phycobilisome rod-core linker polypeptide cpcg2 (28 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- Allophycocyanin subunit alpha 1 (17 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
- C-phycocyanin alpha subunit (17 kDa, Protein from Nostoc sp. PCC 7120 = FACHB-418)
BD23-Fab in complex with the S ectodomain trimer
Resolution: 3.84 Å
EM Method: Single-particle
Fitted PDBs: 7byr
Q-score: 0.441
Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, Zhu Q, Zhang X, Zheng Y, Geng C, Chai X, He R, Li X, Lv Q, Zhu H, Deng W, Xu Y, Wang Y, Qiao L, Tan Y, Song L, Wang G, Du X, Gao N, Liu J, Xiao J, Su XD, Du Z, Feng Y, Qin C, Qin C, Jin R, Xie XS
Cell (2020) 182 pp. 73-84.e16 [ Pubmed: 32425270 DOI: doi:10.1016/j.cell.2020.05.025 ]
- Ab23-fab-heavy chain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike glycoprotein (Complex from SARS coronavirus 2)
- Ab23-fab (Complex from Homo sapiens)
- Bd23-fab in complex with the s ectodomain trimer (Complex)
- Spike glycoprotein (138 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ab23-fab-light chain (11 kDa, Protein from Homo sapiens)
Resolution: 2.37 Å
EM Method: Single-particle
Fitted PDBs: 6k61
Q-score: 0.65
Zheng L, Li Y, Li X, Zhong Q, Li N, Zhang K, Zhang Y, Chu H, Ma C, Li G, Zhao J, Gao N
Nat Plants (2019) 5 pp. 1087-1097 [ DOI: doi:10.1038/s41477-019-0525-6 Pubmed: 31595062 ]
- Photosystem i reaction center subunit viii (5 kDa, Protein from Nostoc sp. PCC 7120)
- Photosystem i reaction center subunit xii (3 kDa, Protein from Nostoc sp. PCC 7120)
- 1,2-di-o-acyl-3-o-[6-deoxy-6-sulfo-alpha-d-glucopyranosyl]-sn-glycerol (795 Da, Ligand)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- Photosystem i reaction center subunit ix (5 kDa, Protein from Nostoc sp. PCC 7120)
- Photosystem i reaction center subunit xi (18 kDa, Protein from Nostoc sp. PCC 7120)
- Photosystem i p700 chlorophyll a apoprotein a1 (83 kDa, Protein from Nostoc sp. PCC 7120)
- Photosystem i 4.8 kda protein (4 kDa, Protein from Nostoc sp. PCC 7120)
- Photosystem i p700 chlorophyll a apoprotein a2 1 (83 kDa, Protein from Nostoc sp. PCC 7120)
- Photosystem i reaction center subunit psak 1 (8 kDa, Protein from Nostoc sp. PCC 7120)
- Photosystem i reaction center subunit iii (17 kDa, Protein from Nostoc sp. PCC 7120)
- Chlorophyll a isomer (893 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Photosystem i reaction center subunit ii (15 kDa, Protein from Nostoc sp. PCC 7120)
- Iron/sulfur cluster (351 Da, Ligand)
- Phylloquinone (450 Da, Ligand)
- Photosystem i reaction center subunit iv (7 kDa, Protein from Nostoc sp. PCC 7120)
- Beta-carotene (536 Da, Ligand)
- Photosystem i complex of cyanobacterium anabaena sp. pcc 7120 (1 MDa, Cellular component from Nostoc sp. PCC 7120)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Photosystem i iron-sulfur center (8 kDa, Protein from Nostoc sp. PCC 7120)
The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 6jyl
Q-score: 0.452
Yan L, Wu H, Li X, Gao N, Chen Z
Nat Struct Mol Biol (2019) 26 pp. 258-266 [ Pubmed: 30872815 DOI: doi:10.1038/s41594-019-0199-9 ]
- Iswi (Complex from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Dna (Complex from Escherichia coli K-12)
- Iswi-nucleosome-crosslinked (300 kDa, Complex)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Iswi chromatin-remodeling complex atpase isw1 (123 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Beryllium trifluoride ion (66 Da, Ligand)
- Nucleosome (Complex from Xenopus laevis)
- Dna (167-mer) (51 kDa, DNA from Escherichia coli K-12)
- Dna (167-mer) (51 kDa, Protein from Escherichia coli K-12)
The complex of ISWI-nucleosome in the ADP.BeF-bound state
Resolution: 3.87 Å
EM Method: Single-particle
Fitted PDBs: 6k1p
Q-score: 0.42
Yan L, Wu H, Li X, Gao N, Chen Z
Nat Struct Mol Biol (2019) 26 pp. 258-266 [ DOI: doi:10.1038/s41594-019-0199-9 Pubmed: 30872815 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Dna (167-mer) (51 kDa, DNA from Escherichia coli K-12)
- Isw1 (Complex from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Nuleosome (Complex from Xenopus laevis)
- Iswi chromatin-remodeling complex atpase isw1 (123 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Dna (167-mer) (51 kDa, DNA from Escherichia coli K-12)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna (Complex from Escherichia coli K-12)
- Beryllium trifluoride ion (66 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- The complex of iswi-nucleosome in the adp-bef-bound state (300 kDa, Complex)
the crosslinked complex of ISWI-nucleosome in the ADP-bound state
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6iro
Q-score: 0.435
Yan L, Wu H, Li X, Gao N, Chen Z
Nat Struct Mol Biol (2019) 26 pp. 258-266 [ Pubmed: 30872815 DOI: doi:10.1038/s41594-019-0199-9 ]
- Dna (167-mer) (51 kDa, DNA from Escherichia coli K-12)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- Iswi chromatin-remodeling complex atpase isw1 (123 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Dna (Complex from Escherichia coli K-12)
- Dna (167-mer) (51 kDa, DNA from Escherichia coli K-12)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- The crosslinked complex of iswi-nucleosome in the adp-bound state (Complex)
- Histone octamer (Complex from Xenopus laevis)
- Isw1 (Complex from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Adenosine-5'-diphosphate (427 Da, Ligand)
Cryo-EM structures of the 50S ribosome subunit bound with HflX
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 5ady
Q-score: 0.289
Zhang Y, Mandava CS, Cao W, Li X, Zhang D, Li N, Zhang Y, Zhang X, Qin Y, Mi K, Lei J, Sanyal S, Gao N
Nat Struct Mol Biol (2015) 22 pp. 906-913 [ Pubmed: 26458047 DOI: doi:10.1038/nsmb.3103 ]
- 50s-hflx complex (1 MDa, Sample)
- Prokaryotic 50s ribosome subunit (1 MDa, Complex from Escherichia coli K-12)
- Hflx (50 kDa, Protein from Escherichia coli)