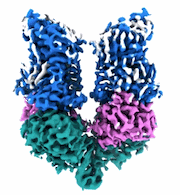
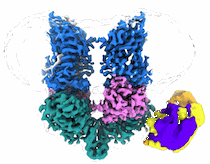Structural Basis of Human NOX5 Activation
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8u85
Q-score: 0.502
Cui C, Jiang M, Jain N, Das S, Lo YH, Kermani AA, Pipatpolkai T, Sun J
Nat Commun (2024) 15 pp. 3994-3994 [ DOI: doi:10.1038/s41467-024-48467-y Pubmed: 38734761 ]
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Nadph oxidase 5 (82 kDa, Protein from Homo sapiens)
- Ala-ala-ala-ala-ala-ala-ala-ala-ala-ala-ala-ala-ala (942 Da, Protein from Homo sapiens)
- Nadph oxidase 5 (Complex from Homo sapiens)
- Nadph dihydro-nicotinamide-adenine-dinucleotide phosphate (745 Da, Ligand)
- Heme b/c (618 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
Structural Basis of Human NOX5 Activation
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8u86
Q-score: 0.45
Cui C, Jiang M, Jain N, Das S, Lo YH, Kermani AA, Pipatpolkai T, Sun J
Nat Commun (2024) 15 pp. 3994-3994 [ DOI: doi:10.1038/s41467-024-48467-y Pubmed: 38734761 ]
- Zinc ion (65 Da, Ligand)
- Nadph oxidase 5 (82 kDa, Protein from Homo sapiens)
- Nadph oxidase 5 (Complex from Homo sapiens)
- Decane (142 Da, Ligand)
- Nadph dihydro-nicotinamide-adenine-dinucleotide phosphate (745 Da, Ligand)
- Heme b/c (618 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Dodecane (170 Da, Ligand)
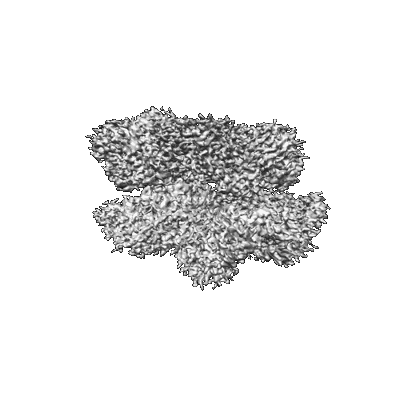
Structural Basis of Human NOX5 Activation
Resolution: 3.86 Å
EM Method: Single-particle
Fitted PDBs: 8u87
Q-score: 0.432
Cui C, Jiang M, Jain N, Das S, Lo YH, Kermani AA, Pipatpolkai T, Sun J
Nat Commun (2024) 15 pp. 3994-3994 [ DOI: doi:10.1038/s41467-024-48467-y Pubmed: 38734761 ]
- Zinc ion (65 Da, Ligand)
- Nadph oxidase 5 (82 kDa, Protein from Homo sapiens)
- Nadph oxidase 5 (Complex from Homo sapiens)
- Nadph dihydro-nicotinamide-adenine-dinucleotide phosphate (745 Da, Ligand)
- Heme b/c (618 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Dodecane (170 Da, Ligand)
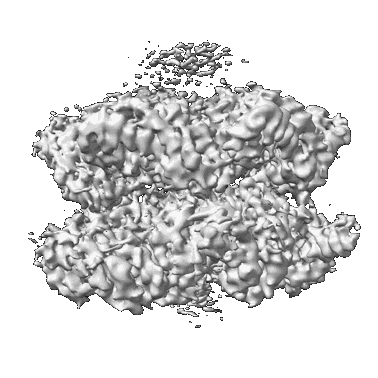
Structural Basis of Human NOX5 Activation
Resolution: 4.06 Å
EM Method: Single-particle
Fitted PDBs: 8u7y
Q-score: 0.327
Cui C, Jiang M, Jain N, Das S, Lo YH, Kermani AA, Pipatpolkai T, Sun J
Nat Commun (2024) 15 pp. 3994-3994 [ DOI: doi:10.1038/s41467-024-48467-y Pubmed: 38734761 ]
- Zinc ion (65 Da, Ligand)
- Nadph oxidase 5 (82 kDa, Protein from Homo sapiens)
- Nadp nicotinamide-adenine-dinucleotide phosphate (743 Da, Ligand)
- Nadph oxidase 5 (Complex from Homo sapiens)
- Decane (142 Da, Ligand)
- Heme b/c (618 Da, Ligand)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Dodecane (170 Da, Ligand)
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8dwu
Q-score: 0.284
Cuneo MJ, O'Flynn BG, Lo YH, Sabri N, Mittag T
Mol Cell (2023) 83 pp. 731-745.e4 [ Pubmed: 36693379 DOI: doi:10.1016/j.molcel.2022.12.033 ]
- Water (18 Da, Ligand)
- Speckle-type poz protein (42 kDa, Protein from Homo sapiens)
- Spop e47k mutant (Complex from Homo sapiens)
Human Kv4.2-KChIP2-DPP6 channel complex in an open state, intracellular region
Resolution: 2.33 Å
EM Method: Single-particle
Fitted PDBs: 7ukh
Q-score: 0.607
Ye W, Zhao H, Dai Y, Wang Y, Lo YH, Jan LY, Lee CH
Mol Cell (2022) 82 pp. 2427- [ Pubmed: 35597238 DOI: doi:10.1016/j.molcel.2022.04.032 ]
- Potassium voltage-gated channel subfamily d member 2 (59 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Isoform 2 of kv channel-interacting protein 2 (28 kDa, Protein from Homo sapiens)
- Human kv4.2-kchip2-dpp6 channel complex in an open state, intracellular region (Complex from Homo sapiens)
CryoEM structure of the N-Terminal deleted Rix7 AAA-ATPase
Resolution: 2.88 Å
EM Method: Single-particle
Fitted PDBs: 7swl
Q-score: 0.546
Kocaman S, Lo YH, Krahn JM, Sobhany M, Dandey VP, Petrovich ML, Etigunta SK, Williams JG, Deterding LJ, Borgnia MJ, Stanley RE
Pnas Nexus (2022) 1 pp. pgac118-pgac118 [ DOI: doi:10.1093/pnasnexus/pgac118 Pubmed: 36090660 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Rix7 (69 kDa, Protein from Chaetomium thermophilum)
- Cryoem structure of the n-terminal-deleted rix7 aaa-atpase (100 kDa, Complex from Chaetomium thermophilum)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Polyleucine (2 kDa, Protein from synthetic construct)
CryoEM structure of the crosslinked Rix7 AAA-ATPase
Resolution: 3.67 Å
EM Method: Single-particle
Fitted PDBs: 7t0v
Q-score: 0.381
Kocaman S, Lo YH, Krahn JM, Sobhany M, Dandey VP, Petrovich ML, Etigunta SK, Williams JG, Deterding LJ, Borgnia MJ, Stanley RE
Pnas Nexus (2022) 1 pp. pgac118-pgac118 [ DOI: doi:10.1093/pnasnexus/pgac118 Pubmed: 36090660 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Cryoem structure of the crosslinked rix7 aaa-atpase (100 kDa, Complex from Chaetomium thermophilum)
- Polyvaline (2 kDa, Protein from synthetic construct)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Rix7 (89 kDa, Protein from Chaetomium thermophilum)
CryoEM structure of the Rix7 D2 Walker B mutant
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 7t3i
Q-score: 0.38
Kocaman S, Lo YH, Krahn JM, Sobhany M, Dandey VP, Petrovich ML, Etigunta SK, Williams JG, Deterding LJ, Borgnia MJ, Stanley RE
Pnas Nexus (2022) 1 pp. pgac118-pgac118 [ DOI: doi:10.1093/pnasnexus/pgac118 Pubmed: 36090660 ]
- Phosphate ion (94 Da, Ligand)
- Substrate peptide (2 kDa, Protein from Escherichia coli)
- Rix7 (89 kDa, Protein from Chaetomium thermophilum)
- Cryoem structure of rix7 d2 walker b mutant (Complex from Chaetomium thermophilum)
- Adenosine-5'-triphosphate (507 Da, Ligand)
Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7
Resolution: 4.5 Å
EM Method: Single-particle
Fitted PDBs: 6mat
Q-score: 0.23
Lo YH, Sobhany M, Hsu AL, Ford BL, Krahn JM, Borgnia MJ, Stanley RE
Nat Commun (2019) 10 pp. 513-513 [ DOI: doi:10.1038/s41467-019-08373-0 Pubmed: 30705282 ]
- Aaa-atpase rix7 (Complex from Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719))
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Unknown protein (2 kDa, Protein from Chaetomium thermophilum var. thermophilum DSM 1495)
- Rix7 mutant (89 kDa, Protein from Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719))