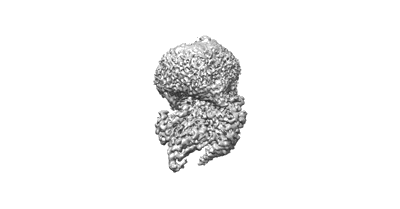
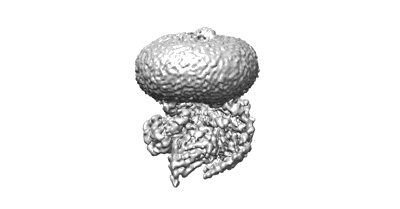
Cryo-EM structure of partial VP4 from simian rotavirus SA11
Resolution: 3.84 Å
EM Method: Single-particle
Fitted PDBs: 8ygr
Q-score: 0.413
Huang Y, Song F, Zeng Y, Sun H, Sheng R, Wang X, Liu L, Luo G, Jiang Y, Chen Y, Zhang M, Zhang S, Gu Y, Yu H, Li S, Li T, Zheng Q, Ge S, Zhang J, Xia N
Nat Commun (2025) 16 pp. 838-838 [ Pubmed: 39833145 DOI: doi:10.1038/s41467-025-56114-3 ]
- Outer capsid protein vp4 (64 kDa, Protein from Rotavirus A)
- Outer capsid protein vp4 (64 kDa, Protein from Rotavirus A)
- Rotavirus a (Virus from Rotavirus A)
- Outer capsid protein vp4 (64 kDa, Protein from Rotavirus A)
- Partial vp4 of rotavirus (Complex from Rotavirus A)
Cryo-EM structure of simian rotavirus SA11 VP4 in complex with nAb 7H13
Resolution: 3.47 Å
EM Method: Single-particle
Fitted PDBs: 8ygs
Q-score: 0.444
Huang Y, Song F, Zeng Y, Sun H, Sheng R, Wang X, Liu L, Luo G, Jiang Y, Chen Y, Zhang M, Zhang S, Gu Y, Yu H, Li S, Li T, Zheng Q, Ge S, Zhang J, Xia N
Nat Commun (2025) 16 pp. 838-838 [ Pubmed: 39833145 DOI: doi:10.1038/s41467-025-56114-3 ]
- Antibody 7h13 heavy chain (12 kDa, Protein from Mus musculus)
- Outer capsid protein vp4 (64 kDa, Protein from Rotavirus A)
- Immune complex of rotavirus with 7h13 (Complex from Rotavirus A)
- Rotavirus vp4 (Complex from Rotavirus A)
- Outer capsid protein vp4 (64 kDa, Protein from Rotavirus A)
- Fab 7h13 (Complex from Mus musculus)
- Antibody 7h13 light chain (11 kDa, Protein from Mus musculus)
Cryo-EM structure of simian rotavirus SA11 VP4 in complex with nAb 7H13 at Low concentration
Cryo-EM structure of simian rotavirus SA11 VP4 in complex with nAb 7H13-I54G mutant (left side)
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 8ygt
Q-score: 0.559
Huang Y, Song F, Zeng Y, Sun H, Sheng R, Wang X, Liu L, Luo G, Jiang Y, Chen Y, Zhang M, Zhang S, Gu Y, Yu H, Li S, Li T, Zheng Q, Ge S, Zhang J, Xia N
Nat Commun (2025) 16 pp. 838-838 [ Pubmed: 39833145 DOI: doi:10.1038/s41467-025-56114-3 ]
- Antibody 7h13-i54g mutant light chain (11 kDa, Protein from Mus musculus)
- Immune complex of rotavirus with 7h13 (Complex from Rotavirus A)
- Outer capsid protein vp4 (64 kDa, Protein from Rotavirus A)
- Rotavirus vp4 (Complex from Rotavirus A)
- Fab 7h13 (Complex from Mus musculus)
- Antibody 7h13-i54g mutant heavy chain (12 kDa, Protein from Mus musculus)
Cryo-EM structure of simian rotavirus SA11 VP4 in complex with nAb 7H13-I54G mutant (right side)
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 8ygu
Q-score: 0.526
Huang Y, Song F, Zeng Y, Sun H, Sheng R, Wang X, Liu L, Luo G, Jiang Y, Chen Y, Zhang M, Zhang S, Gu Y, Yu H, Li S, Li T, Zheng Q, Ge S, Zhang J, Xia N
Nat Commun (2025) 16 pp. 838-838 [ Pubmed: 39833145 DOI: doi:10.1038/s41467-025-56114-3 ]
- Outer capsid protein vp4 (87 kDa, Protein from Rotavirus A)
- Outer capsid protein vp4 (64 kDa, Protein from Rotavirus A)
- Antibody 7h13-i54g mutant light chain (11 kDa, Protein from Mus musculus)
- Fab 7h13 (Complex from Rotavirus A)
- Antibody 7h13-i54g mutant heavy chain (12 kDa, Protein from Mus musculus)
- Rotavirus vp4 (Complex from Rotavirus A)
- Immune complex of rotavirus with 7h13-i54g mutant (Complex from Rotavirus A)
Cryo-EM structure of peptide free and Gs-coupled GIPR
Resolution: 2.86 Å
EM Method: Single-particle
Fitted PDBs: 8wa3
Q-score: 0.464
Cong Z, Zhao F, Li Y, Luo G, Mai Y, Chen X, Chen Y, Lin S, Cai X, Zhou Q, Yang D, Wang MW
Cell Discov (2024) 10 pp. 18-18 [ DOI: doi:10.1038/s41421-024-00649-0 Pubmed: 38346960 ]
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1,o-antigen polymerase (40 kDa, Protein from Leptolinea tardivitalis)
- Nanobody-35 (15 kDa, Protein from synthetic construct)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Bos taurus)
- Cryo-em structure of the human glucose-dependent insulinotropic polypeptide receptor in complex with gip and g protein (Complex from Homo sapiens)
- Gastric inhibitory polypeptide receptor,fusion protein (64 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(s) subunit alpha isoforms short (45 kDa, Protein from Bos taurus)
Cryo-EM structures of peptide free and Gs-coupled GLP-1R
Resolution: 2.54 Å
EM Method: Single-particle
Fitted PDBs: 8wg7
Q-score: 0.513
Cong Z, Zhao F, Li Y, Luo G, Mai Y, Chen X, Chen Y, Lin S, Cai X, Zhou Q, Yang D, Wang MW
Cell Discov (2024) 10 pp. 18-18 [ DOI: doi:10.1038/s41421-024-00649-0 Pubmed: 38346960 ]
- Nanobody-35 (13 kDa, Protein from synthetic construct)
- Guanine nucleotide-binding protein g(s) subunit alpha isoforms short (45 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (40 kDa, Protein from Rattus norvegicus)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Bos taurus)
- Cryo-em structure of the human glucagon like peptide 1 receptor in complex with tirzepatide and g protein (Complex from Homo sapiens)
- Glucagon-like peptide 1 receptor (50 kDa, Protein from Homo sapiens)
Cryo-EM structures of peptide free and Gs-coupled GCGR
Resolution: 2.71 Å
EM Method: Single-particle
Fitted PDBs: 8wg8
Q-score: 0.498
Cong Z, Zhao F, Li Y, Luo G, Mai Y, Chen X, Chen Y, Lin S, Cai X, Zhou Q, Yang D, Wang MW
Cell Discov (2024) 10 pp. 18-18 [ DOI: doi:10.1038/s41421-024-00649-0 Pubmed: 38346960 ]
- Guanine nucleotide-binding protein g(s) subunit alpha isoforms short (41 kDa, Protein from Homo sapiens)
- Nanobody-35 (15 kDa, Protein from synthetic construct)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (40 kDa, Protein from Rattus norvegicus)
- Pro-pro-pro-pro-phe-ser-asn-leu-val-met-asp-asp-leu-lys-asn-lys-lys (1 kDa, Protein from Spodoptera frugiperda)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Bos taurus)
- Cryo-em structure of the human glucagon like peptide 1 receptor in complex with tirzepatide and g protein (Complex from Homo sapiens)
- Glucagon receptor (46 kDa, Protein from Homo sapiens)
Resolution: 2.12 Å
EM Method: Single-particle
Fitted PDBs: 7xy9
Q-score: 0.714
Chen Q, Qu G, Li X, Feng M, Yang F, Li Y, Li J, Tong F, Song S, Wang Y, Sun Z, Luo G
Nat Commun (2023) 14 pp. 2117-2117 [ Pubmed: 37055470 DOI: doi:10.1038/s41467-023-37921-y ]
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Enzyme assembled gel (Complex from Thermoanaerobacter brockii)
- Nadp-dependent isopropanol dehydrogenase (38 kDa, Protein from Thermoanaerobacter brockii)
Cryo-EM Structure of the Exocyst Complex
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 5yfp
Q-score: 0.155
Mei K, Li Y, Wang S, Shao G, Wang J, Ding Y, Luo G, Yue P, Liu JJ, Wang X, Dong MQ, Wang HW, Guo W
Nat Struct Mol Biol (2018) 25 pp. 139-146 [ Pubmed: 29335562 DOI: doi:10.1038/s41594-017-0016-2 ]
- Exocyst complex component sec10 (100 kDa, Protein from Saccharomyces cerevisia S288c)
- Exocyst complex component exo84 (85 kDa, Protein from Saccharomyces cerevisiae S288c)
- Exocyst complex component sec5 (112 kDa, Protein from Saccharomyces cerevisia S288c)
- Exocyst complex component sec6 (93 kDa, Protein from Saccharomyces cerevisia S288c)
- Exocyst complex component sec8 (122 kDa, Protein from Saccharomyces cerevisia S288c)
- Exocyst complex component exo70 (71 kDa, Protein from Saccharomyces cerevisia S288c)
- Cryo-em map of the yeast exocyst complex overall structure at 5.5a resolution (844 kDa, Complex from Saccharomyces cerevisia S288c)
- Exocyst complex component sec15 (105 kDa, Protein from Saccharomyces cerevisia S288c)
- Exocyst complex component sec3 (154 kDa, Protein from Saccharomyces cerevisiae S288c)