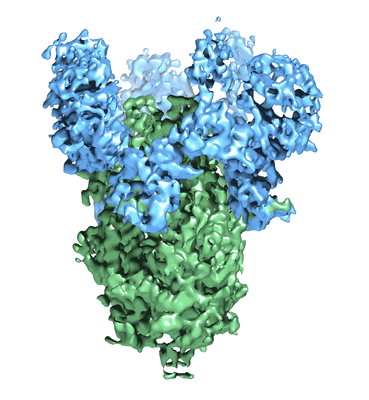
Negative stain EM map of MERS 2 in complex with G2 Fab
Wang N, Rosen O, Wang L, Turner HL, Stevens LJ, Corbett KS, Bowman CA, Pallesen J, Shi W, Zhang Y, Leung K, Kirchdoerfer RN, Becker MM, Denison MR, Chappell JD, Ward AB, Graham BS, McLellan JS
Cell Rep (2019) 28 pp. 3395-3405.e6 [ Pubmed: 31553909 DOI: doi:10.1016/j.celrep.2019.08.052 ]
MERS S0 trimer in complex with antibody G2
Resolution: 4.19 Å
EM Method: Single-particle
Fitted PDBs: 6pz8
Q-score: 0.133
Wang N, Rosen O, Wang L, Turner HL, Stevens LJ, Corbett KS, Bowman CA, Pallesen J, Shi W, Zhang Y, Leung K, Kirchdoerfer RN, Becker MM, Denison MR, Chappell JD, Ward AB, Graham BS, McLellan JS
Cell Rep (2019) 28 pp. 3395-3405.e6 [ Pubmed: 31553909 DOI: doi:10.1016/j.celrep.2019.08.052 ]
- G2 light chain (23 kDa, Protein from Mus musculus)
- Antibody g2 (Complex from Mus musculus)
- S protein (51 kDa, Protein from Middle East respiratory syndrome-related coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Mers s0 ectodomain trimer in complex with antibody g2 (Complex)
- G2 heavy chain (24 kDa, Protein from Mus musculus)
- Mers s0 ectodomain trimer (Complex from Middle East respiratory syndrome-related coronavirus)
- S protein (80 kDa, Protein from Middle East respiratory syndrome-related coronavirus)
SARS Spike Glycoprotein, Stabilized variant, C3 symmetry
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 6crv
Q-score: 0.466
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ Pubmed: 30356097 DOI: doi:10.1038/s41598-018-34171-7 ]
- Spike glycoprotein,fibritin (137 kDa, Protein from Enterobacteria phage T4)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars spike glycoprotein, stabilized ectodomain (540 kDa, Complex from SARS coronavirus)
SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 6crw
Q-score: 0.283
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ DOI: doi:10.1038/s41598-018-34171-7 Pubmed: 30356097 ]
- Spike glycoprotein,fibritin (134 kDa, Protein from Enterobacteria phage T4)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars spike glycoprotein, stabilized variant (540 kDa, Complex from SARS coronavirus)
SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 6crx
Q-score: 0.324
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ DOI: doi:10.1038/s41598-018-34171-7 Pubmed: 30356097 ]
- Spike glycoprotein,fibritin (134 kDa, Protein from Enterobacteria phage T4)
- Sars spike glycoprotein (540 kDa, Complex from SARS coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS spike glycoprotein, stabilized variant, three S1 CTDs in an upwards conformation
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ DOI: doi:10.1038/s41598-018-34171-7 Pubmed: 30356097 ]
- Sars spike glycoprotein (540 kDa, Complex from SARS coronavirus)
- Sars spike glycoprotein (Protein from SARS coronavirus)
SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, C3 symmetry
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 6crz
Q-score: 0.012
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ Pubmed: 30356097 DOI: doi:10.1038/s41598-018-34171-7 ]
- Sars spike glycoprotein,trypsin-cleaved, stabilized variant (540 kDa, Complex from SARS coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein,fibritin (134 kDa, Protein from Enterobacteria phage T4)
SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, one S1 CTD in an upwards conformation
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 6cs0
Q-score: 0.275
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ DOI: doi:10.1038/s41598-018-34171-7 Pubmed: 30356097 ]
- Sars spike glycoprotein,trypsin-cleaved, stabilized variant (540 kDa, Complex from SARS coronavirus)
- Spike glycoprotein,fibritin (134 kDa, Protein from Enterobacteria phage T4)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation
Resolution: 4.6 Å
EM Method: Single-particle
Fitted PDBs: 6cs1
Q-score: 0.041
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ Pubmed: 30356097 DOI: doi:10.1038/s41598-018-34171-7 ]
- Sars spike glycoprotein,trypsin-cleaved, stabilized variant (540 kDa, Complex from SARS coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein,fibritin (134 kDa, Protein from Enterobacteria phage T4)
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB
Sci Rep (2018) 8 pp. 15701-15701 [ Pubmed: 30356097 DOI: doi:10.1038/s41598-018-34171-7 ]