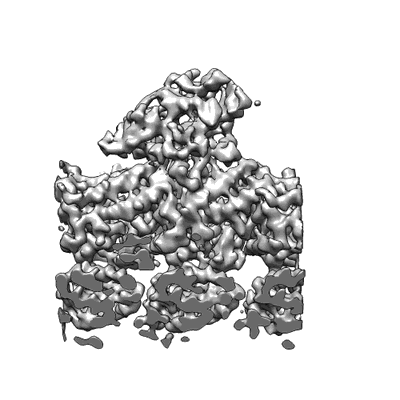
SARS CoV-2 Spike protein, Closed conformation, C1 symmetry
Resolution: 3.03 Å
EM Method: Single-particle
Fitted PDBs: 6zb4
Q-score: 0.587
Toelzer C, Gupta K, Yadav SKN, Borucu U, Davidson AD, Kavanagh Williamson M, Shoemark DK, Garzoni F, Staufer O, Milligan R, Capin J, Mulholland AJ, Spatz J, Fitzgerald D, Berger I, Schaffitzel C
Science (2020) 370 pp. 725-730 [ Pubmed: 32958580 DOI: doi:10.1126/science.abd3255 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Linoleic acid (280 Da, Ligand)
- Spike glycoprotein (139 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars cov-2 spike protein, closed conformation, c1 symmetry (540 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
SARS CoV-2 Spike protein, Closed conformation, C3 symmetry
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 6zb5
Q-score: 0.619
Toelzer C, Gupta K, Yadav SKN, Borucu U, Davidson AD, Kavanagh Williamson M, Shoemark DK, Garzoni F, Staufer O, Milligan R, Capin J, Mulholland AJ, Spatz J, Fitzgerald D, Berger I, Schaffitzel C
Science (2020) 370 pp. 725-730 [ Pubmed: 32958580 DOI: doi:10.1126/science.abd3255 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars cov-2 spike protein, closed conformation, c3 symmetry (540 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Linoleic acid (280 Da, Ligand)
- Spike glycoprotein (139 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS CoV-2 Spike protein, Open conformation
Toelzer C, Gupta K, Yadav SKN, Borucu U, Davidson AD, Kavanagh Williamson M, Shoemark DK, Garzoni F, Staufer O, Milligan R, Capin J, Mulholland AJ, Spatz J, Fitzgerald D, Berger I, Schaffitzel C
Science (2020) 370 pp. 725-730 [ Pubmed: 32958580 DOI: doi:10.1126/science.abd3255 ]
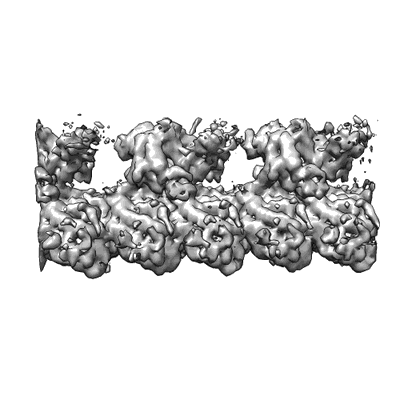
Cryo-EM structure of microtubule-bound KLP61F motor domain in the AMPPNP state
Resolution: 4.4 Å
EM Method: Helical reconstruction
Fitted PDBs: 6vpo
Q-score: 0.324
Bodrug T, Wilson-Kubalek EM, Nithianantham S, Thompson AF, Alfieri A, Gaska I, Major J, Debs G, Inagaki S, Gutierrez P, Gheber L, McKenney RJ, Sindelar CV, Milligan R, Stumpff J, Rosenfeld SS, Forth ST, Al-Bassam J
eLife (2020) 9 [ DOI: doi:10.7554/eLife.51131 Pubmed: 31958056 ]
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Tubulin beta chain (49 kDa, Protein from Sus scrofa)
- Tubulin alpha-1a chain (50 kDa, Protein from Sus scrofa)
- Microtubule (Complex from Sus scrofa)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Kinesin-like protein klp61f (42 kDa, Protein from Drosophila melanogaster)
- Microtubule-bound klp61f motor in the amppnp state (Complex)
- Klp61f motor (Complex from Drosophila melanogaster)
Cryo-EM structure of microtubule-bound KLP61F motor with tail domain in the nucleotide-free state
Resolution: 4.4 Å
EM Method: Helical reconstruction
Fitted PDBs: 6vpp
Q-score: 0.218
Bodrug T, Wilson-Kubalek EM, Nithianantham S, Thompson AF, Alfieri A, Gaska I, Major J, Debs G, Inagaki S, Gutierrez P, Gheber L, McKenney RJ, Sindelar CV, Milligan R, Stumpff J, Rosenfeld SS, Forth ST, Al-Bassam J
eLife (2020) 9 [ DOI: doi:10.7554/eLife.51131 Pubmed: 31958056 ]
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Tubulin beta chain (49 kDa, Protein from Sus scrofa)
- Tubulin alpha-1a chain (50 kDa, Protein from Sus scrofa)
- Microtubule (Complex from Sus scrofa)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Kinesin-like protein klp61f (42 kDa, Protein from Drosophila melanogaster)
- Microtubule-bound klp61f motor with tail domain in the nucleotide-free state (Complex)
- Klp61f motor (Complex from Drosophila melanogaster)