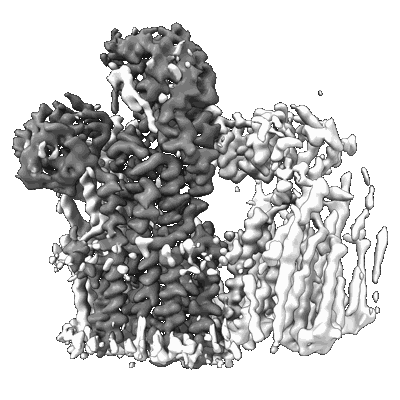
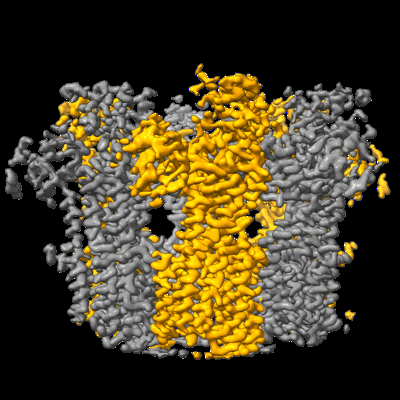
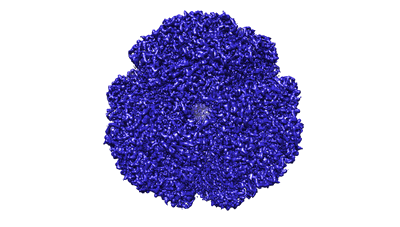
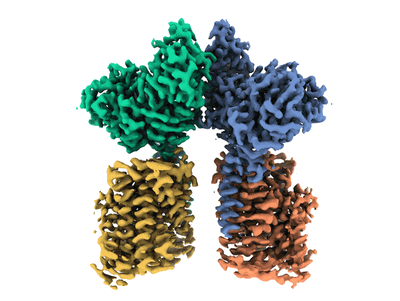
Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7nxf
Q-score: 0.439
Heit S, Geurts MMG, Murphy BJ, Corey RA, Mills DJ, Kuhlbrandt W, Bublitz M
Sci Adv (2021) 7 pp. eabj5255-eabj5255 [ DOI: doi:10.1126/sciadv.abj5255 Pubmed: 34757782 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Potassium ion (39 Da, Ligand)
- Monomeric subunit of the hexameric fungal plasma membrane proton pump in its auto-inhibited state (Cellular component from Neurospora crassa)
- Magnesium ion (24 Da, Ligand)
- Plasma membrane atpase (99 kDa, Protein from Neurospora crassa)
Resolution: 3.26 Å
EM Method: Single-particle
Fitted PDBs: 7ny1
Q-score: 0.357
Heit S, Geurts MMG, Murphy BJ, Corey RA, Mills DJ, Kuhlbrandt W, Bublitz M
Sci Adv (2021) 7 pp. eabj5255-eabj5255 [ DOI: doi:10.1126/sciadv.abj5255 Pubmed: 34757782 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Potassium ion (39 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Plasma membrane atpase (99 kDa, Protein from Neurospora crassa)
- Hexameric assembly of the fungal plasma membrane proton pump in its auto-inhibited state (Complex from Neurospora crassa)
Helicobacter pylori urease with BME bound in the active site
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 6qsu
Q-score: 0.656
Cunha ES, Chen X, Sanz-Gaitero M, Mills DJ, Luecke H
Nat Commun (2021) 12 pp. 230-230 [ Pubmed: 33431861 DOI: doi:10.1038/s41467-020-20485-6 ]
- Nickel (ii) ion (58 Da, Ligand)
- 1.1 mda helicobacter pylori urease (1 MDa, Complex from Helicobacter pylori)
- Urease subunit alpha (26 kDa, Protein from Helicobacter pylori)
- Beta-mercaptoethanol (78 Da, Ligand)
- Urease subunit beta (61 kDa, Protein from Helicobacter pylori)
- Water (18 Da, Ligand)
Heterotetrameric structure of the rBAT-b(0,+)AT1 complex
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6yup
Q-score: 0.491
Wu D, Grund TN, Welsch S, Mills DJ, Michel M, Safarian S, Michel H
PNAS (2020) 117 pp. 21281-21287 [ DOI: doi:10.1073/pnas.2008111117 Pubmed: 32817565 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Neutral and basic amino acid transport protein rbat (78 kDa, Protein from Homo sapiens)
- Heterotetrameric complex of rbat and b(0,+)at1 (264 MDa, Complex from Homo sapiens)
- B(0,+)-type amino acid transporter 1 (53 kDa, Protein from Homo sapiens)
Homodimeric structure of the rBAT complex
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 6yuz
Q-score: 0.56
Wu D, Grund TN, Welsch S, Mills DJ, Michel M, Safarian S, Michel H
PNAS (2020) 117 pp. 21281-21287 [ DOI: doi:10.1073/pnas.2008111117 Pubmed: 32817565 ]
- Neutral and basic amino acid transport protein rbat (78 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Homodimeric complex of rbat (157 MDa, Complex from Homo sapiens)
- Calcium ion (40 Da, Ligand)
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6yv1
Q-score: 0.447
Wu D, Grund TN, Welsch S, Mills DJ, Michel M, Safarian S, Michel H
PNAS (2020) 117 pp. 21281-21287 [ DOI: doi:10.1073/pnas.2008111117 Pubmed: 32817565 ]
- Human b(0,+)at1 (106 MDa, Complex from Homo sapiens)
- B(0,+)-type amino acid transporter 1 (53 kDa, Protein from Homo sapiens)
Helicobacter pylori urease with inhibitor bound in the active site
Resolution: 2.0 Å
EM Method: Single-particle
Fitted PDBs: 6zja
Q-score: 0.758
Cunha ES, Chen X, Sanz-Gaitero M, Mills DJ, Luecke H
Nat Commun (2021) 12 pp. 230-230 [ Pubmed: 33431861 DOI: doi:10.1038/s41467-020-20485-6 ]
- Nickel (ii) ion (58 Da, Ligand)
- 1.1 mda helicobacter pylori urease (1 MDa, Complex from Helicobacter pylori)
- Urease subunit alpha (26 kDa, Protein from Helicobacter pylori)
- Urease subunit beta (61 kDa, Protein from Helicobacter pylori)
- 2-{[1-(3,5-dimethylphenyl)-1h-imidazol-2-yl]sulfanyl}-n-hydroxyacetamide (277 Da, Ligand)
- Water (18 Da, Ligand)
stimulatory human GTP cyclohydrolase I - GFRP complex
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 6z80
Q-score: 0.528
Ebenhoch R, Prinz S, Kaltwasser S, Mills DJ, Meinecke R, Rubbelke M, Reinert D, Bauer M, Weixler L, Zeeb M, Vonck J, Nar H
PNAS (2020) 117 pp. 31838-31849 [ Pubmed: 33229582 DOI: doi:10.1073/pnas.2013473117 ]
- Zinc ion (65 Da, Ligand)
- Gtp cyclohydrolase 1 feedback regulatory protein (9 kDa, Protein from Homo sapiens)
- Stimulatory gch1-gfrp complex (phenylalanine and 8-oxo-gtp bound) (353 kDa, Complex from Homo sapiens)
- Gtp cyclohydrolase 1 (25 kDa, Protein from Homo sapiens)
- Phenylalanine (165 Da, Ligand)
- 8-oxo-guanosine-5'-triphosphate (539 Da, Ligand)
inhibitory human GTP cyclohydrolase I - GFRP complex
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6z85
Q-score: 0.529
Ebenhoch R, Prinz S, Kaltwasser S, Mills DJ, Meinecke R, Rubbelke M, Reinert D, Bauer M, Weixler L, Zeeb M, Vonck J, Nar H
PNAS (2020) 117 pp. 31838-31849 [ Pubmed: 33229582 DOI: doi:10.1073/pnas.2013473117 ]
- Zinc ion (65 Da, Ligand)
- 7,8-dihydrobiopterin (239 Da, Ligand)
- Gtp cyclohydrolase 1 feedback regulatory protein (9 kDa, Protein from Homo sapiens)
- Gtp cyclohydrolase 1 (25 kDa, Protein from Homo sapiens)
- Inhibitory gch1-gfrp complex (bh4 bound) (353 kDa, Complex from Homo sapiens)
Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 6rko
Q-score: 0.665
Safarian S, Hahn A, Mills DJ, Radloff M, Eisinger ML, Nikolaev A, Meier-Credo J, Melin F, Miyoshi H, Gennis RB, Sakamoto J, Langer JD, Hellwig P, Kuhlbrandt W, Michel H
Science (2019) 366 pp. 100-104 [ Pubmed: 31604309 DOI: doi:10.1126/science.aay0967 ]
- Ubiquinone-8 (727 Da, Ligand)
- Cis-heme d hydroxychlorin gamma-spirolactone (632 Da, Ligand)
- Uncharacterized protein ynhf (2 kDa, Protein from Escherichia coli (strain K12))
- Cytochrome bd-i oxidase from e. coli (107 kDa, Complex from Escherichia coli K-12)
- Cytochrome bd-i ubiquinol oxidase subunit 1 (58 kDa, Protein from Escherichia coli (strain K12))
- Cytochrome bd-i ubiquinol oxidase subunit x (4 kDa, Protein from Escherichia coli (strain K12))
- Cytochrome bd-i ubiquinol oxidase subunit 2 (42 kDa, Protein from Escherichia coli (strain K12))
- Heme b/c (618 Da, Ligand)
- Water (18 Da, Ligand)
- Oxygen molecule (31 Da, Ligand)
- (2s)-3-(hexadecanoyloxy)-2-[(9z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate (760 Da, Ligand)