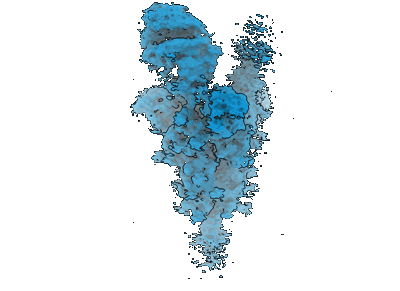
SARS-CoV-2 BA-2.87.1 Spike ectodomain
Duyvesteyn HME, Dijokaite-Guraliuc A, Liu C, Supasa P, Kronsteiner B, Jeffery K, Stafford L, Klenerman P, Dunachie SJ, Mongkolsapaya J, Fry EE, Ren J, Stuart DI, Screaton GR
Structure (2024) 32 pp. 1594- [ Pubmed: 39173622 DOI: doi:10.1016/j.str.2024.07.020 ]
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 8qsq
Q-score: 0.14
Liu C, Zhou D, Dijokaite-Guraliuc A, Supasa P, Duyvesteyn HME, Ginn HM, Selvaraj M, Mentzer AJ, Das R, de Silva TI, Ritter TG, Plowright M, Newman TAH, Stafford L, Kronsteiner B, Temperton N, Lui Y, Fellermeyer M, Goulder P, Klenerman P, Dunachie SJ, Barton MI, Kutuzov MA, Dushek O, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Cell Rep Med (2024) 5 pp. 101553-101553 [ DOI: doi:10.1016/j.xcrm.2024.101553 Pubmed: 38723626 ]
- Spike protein s2' (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba-2.86 variant sars-cov2 s protein complexed with angiotensin converting enzyme 2 (ace2) (Complex)
- Processed angiotensin-converting enzyme 2 (69 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ba-2.86 variant sars-cov2 s protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Angiotensin converting enzyme 2 (ace2) (Complex from Homo sapiens)
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8qtd
Q-score: 0.454
Liu C, Zhou D, Dijokaite-Guraliuc A, Supasa P, Duyvesteyn HME, Ginn HM, Selvaraj M, Mentzer AJ, Das R, de Silva TI, Ritter TG, Plowright M, Newman TAH, Stafford L, Kronsteiner B, Temperton N, Lui Y, Fellermeyer M, Goulder P, Klenerman P, Dunachie SJ, Barton MI, Kutuzov MA, Dushek O, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Cell Rep Med (2024) 5 pp. 101553-101553 [ DOI: doi:10.1016/j.xcrm.2024.101553 Pubmed: 38723626 ]
- Xbb-7 fab light chain (10 kDa, Protein from Homo sapiens)
- Xbb-7 fab heavy chain (14 kDa, Protein from Homo sapiens)
- Ba.2.86 variant sars-cov2 s protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Xbb-7 fab (Complex from Homo sapiens)
- Ba-2.86 variant sars-cov2 s protein complexed with xbb-7 fab. (Complex)
- Spike glycoprotein,fibritin (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Resolution: 3.41 Å
EM Method: Single-particle
Fitted PDBs: 8r8k
Q-score: 0.504
Liu C, Zhou D, Dijokaite-Guraliuc A, Supasa P, Duyvesteyn HME, Ginn HM, Selvaraj M, Mentzer AJ, Das R, de Silva TI, Ritter TG, Plowright M, Newman TAH, Stafford L, Kronsteiner B, Temperton N, Lui Y, Fellermeyer M, Goulder P, Klenerman P, Dunachie SJ, Barton MI, Kutuzov MA, Dushek O, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Cell Rep Med (2024) 5 pp. 101553-101553 [ DOI: doi:10.1016/j.xcrm.2024.101553 Pubmed: 38723626 ]
- Xbb-4 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Xbb-4 fab in complex with sars-cov-2 ba.2.12.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Xbb-4 fab light chain (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein,fibritin (142 kDa, Protein from Tequatrovirus T4)
- Xbb-4 fab (Complex from Homo sapiens)
- Ba.2.12.1 spike glycoprotein ectodomain (Complex from Severe acute respiratory syndrome coronavirus 2)
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
Resolution: 2.2 Å
EM Method: Single-particle
Fitted PDBs: 8r1c
Q-score: 0.645
Zhou D, Supasa P, Liu C, Dijokaite-Guraliuc A, Duyvesteyn HME, Selvaraj M, Mentzer AJ, Das R, Dejnirattisai W, Temperton N, Klenerman P, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Nat Commun (2024) 15 pp. 2734-2734 [ Pubmed: 38548763 DOI: doi:10.1038/s41467-024-46982-6 ]
- Spike glycoprotein,fibritin (142 kDa, Protein from Enterobacteria phage T4)
- Sd1-2 fab heavy chain (13 kDa, Protein from Homo sapiens)
- Sd1-2 fab light chain (11 kDa, Protein from Homo sapiens)
- Sd1-2 fab (Complex from Homo sapiens)
- Ba.2.12.1 spike glycoprotein ectodomain (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sd1-2 fab in complex with sars-cov-2 ba.2.12.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Resolution: 2.37 Å
EM Method: Single-particle
Fitted PDBs: 8r1d
Q-score: 0.658
Zhou D, Supasa P, Liu C, Dijokaite-Guraliuc A, Duyvesteyn HME, Selvaraj M, Mentzer AJ, Das R, Dejnirattisai W, Temperton N, Klenerman P, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Nat Commun (2024) 15 pp. 2734-2734 [ Pubmed: 38548763 DOI: doi:10.1038/s41467-024-46982-6 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sd1-3 fab light chain (11 kDa, Protein from Homo sapiens)
- Sd1-3 fab in complex with sars-cov-2 ba.12.1 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sd1-3 fab heavy chain (12 kDa, Protein from Homo sapiens)
- Spike glycoprotein,fibritin (142 kDa, Protein from Enterobacteria phage T4)
- Sd1-3 fab (Complex from Homo sapiens)
- Water (18 Da, Ligand)
- Ba.2.12.1 spike glycoprotein ectodomain (Complex from Severe acute respiratory syndrome coronavirus 2)
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8cin
Q-score: 0.514
Zhou D, Supasa P, Liu C, Dijokaite-Guraliuc A, Duyvesteyn HME, Selvaraj M, Mentzer AJ, Das R, Dejnirattisai W, Temperton N, Klenerman P, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Nat Commun (2024) 15 pp. 2734-2734 [ Pubmed: 38548763 DOI: doi:10.1038/s41467-024-46982-6 ]
- Ba.4/5-5 fab heavy and light chains. (Complex from Homo sapiens)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ba.4/5-5 fab heavy chain (23 kDa, Protein from Homo sapiens)
- Sars-coronavirus-2 ba.4 spike glycoprotein in complex with ba.4/5-5 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-coronavirus-2 ba.4 spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ba.4/5-5 fab light chain (22 kDa, Protein from Homo sapiens)
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8bcz
Q-score: 0.564
Dijokaite-Guraliuc A, Das R, Zhou D, Ginn HM, Liu C, Duyvesteyn HME, Huo J, Nutalai R, Supasa P, Selvaraj M, de Silva TI, Plowright M, Newman TAH, Hornsby H, Mentzer AJ, Skelly D, Ritter TG, Temperton N, Klenerman P, Barnes E, Dunachie SJ, Roemer C, Peacock TP, Paterson NG, Williams MA, Hall DR, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR
Cell Rep (2023) 42 pp. 112271-112271 [ DOI: doi:10.1016/j.celrep.2023.112271 Pubmed: 36995936 ]
- Complex of sars-cov2 delta variant receptor binding domain with 4 fabs (Complex)
- Ba.2-36 heavy chain (13 kDa, Protein from Homo sapiens)
- Covox-45 heavy chain (24 kDa, Protein from Homo sapiens)
- Antibody chains (Complex from Homo sapiens)
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ba.2-23 light chain (11 kDa, Protein from Homo sapiens)
- Covox-45 light chain (11 kDa, Protein from Homo sapiens)
- Ba.2-36 light chain (11 kDa, Protein from Homo sapiens)
- Sars-cov2 delta variant receptor binding domain (Complex from Severe acute respiratory syndrome coronavirus 2)
- Ey6a heavy chain (13 kDa, Protein from Homo sapiens)
- Ey6a light chain (11 kDa, Protein from Homo sapiens)
- Ba.2-23 heavy chain (12 kDa, Protein from Homo sapiens)
CryoEM structure of sNS1 complexed with Fab5E3
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7wur
Q-score: 0.427
Shu B, Ooi JSG, Tan AWK, Ng TS, Dejnirattisai W, Mongkolsapaya J, Fibriansah G, Shi J, Kostyuchenko VA, Screaton GR, Lok SM
Nat Commun (2022) 13 pp. 6756-6756 [ Pubmed: 36347841 DOI: doi:10.1038/s41467-022-34415-1 ]
- Fab 5e3 (Complex)
- Structure of dengue ns1 complexed with fab 5e3 (Complex from Dengue virus 2)
- Core protein (38 kDa, Protein from Dengue virus 2)
- Dengue ns1 (Complex from Homo sapiens)
- Fab 5e3 light chain (11 kDa, Protein from Homo sapiens)
- Fab 5e3 heavy chain (12 kDa, Protein from Homo sapiens)
CryoEM structure of a dimer of loose sNS1 tetramer
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7wus
Q-score: 0.402
Shu B, Ooi JSG, Tan AWK, Ng TS, Dejnirattisai W, Mongkolsapaya J, Fibriansah G, Shi J, Kostyuchenko VA, Screaton GR, Lok SM
Nat Commun (2022) 13 pp. 6756-6756 [ Pubmed: 36347841 DOI: doi:10.1038/s41467-022-34415-1 ]
- Dengue ns1 dimer of loose tetramer (Cellular component from Dengue virus 2)
- Core protein (38 kDa, Protein from Dengue virus 2)