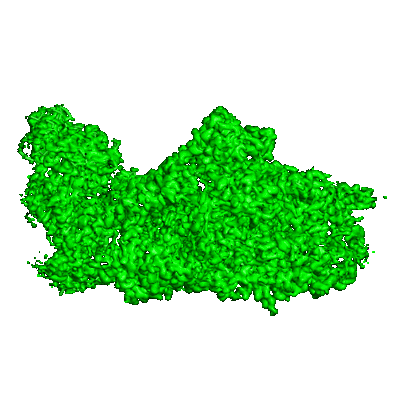
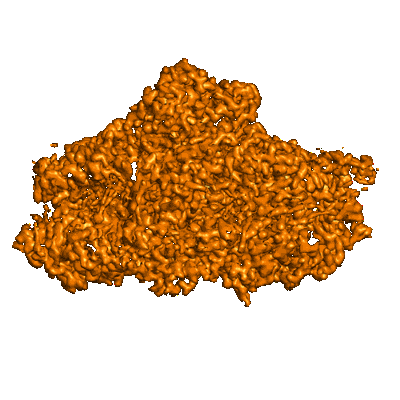
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8am5
Q-score: 0.52
Zhao Z, Vercellino I, Knoppova J, Sobotka R, Murray JW, Nixon PJ, Sazanov LA, Komenda J
Nat Commun (2023) 14 pp. 4681-4681 [ DOI: doi:10.1038/s41467-023-40388-6 Pubmed: 37542031 ]
- Photosystem ii protein d1 2 (38 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit psak 1 (8 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit xii (3 kDa, Protein from Synechocystis sp. PCC 6803)
- Phylloquinone (450 Da, Ligand)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- Beta,beta-caroten-4-one (550 Da, Ligand)
- Photosystem i reaction center subunit iv (8 kDa, Protein from Synechocystis sp. PCC 6803)
- Beta-carotene (536 Da, Ligand)
- Photosystem i reaction center subunit xi (16 kDa, Protein from Synechocystis sp. PCC 6803)
- Rcii/psi complex (Complex from Synechocystis sp. PCC 6803)
- Fe (ii) ion (55 Da, Ligand)
- (1r,2s)-4-{(1e,3e,5e,7e,9e,11e,13e,15e,17e)-18-[(4s)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol (568 Da, Ligand)
- Cytochrome b559 subunit alpha (9 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit ix (4 kDa, Protein from Synechocystis sp. PCC 6803)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Photosystem ii d2 protein (37 kDa, Protein from Synechocystis sp. PCC 6803)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Photosystem i reaction center subunit viii (4 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit ii (15 kDa, Protein from Synechocystis sp. PCC 6803)
- Pheophytin a (871 Da, Ligand)
- Chlorophyll a isomer (893 Da, Ligand)
- Cytochrome b559 subunit beta (4 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i p700 chlorophyll a apoprotein a2 (81 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i iron-sulfur center (8 kDa, Protein from Synechocystis sp. PCC 6803)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- (3'r)-3'-hydroxy-beta,beta-caroten-4-one (566 Da, Ligand)
- Photosystem ii assembly lipoprotein ycf48 (37 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i p700 chlorophyll a apoprotein a1 (83 kDa, Protein from Synechocystis sp. PCC 6803)
- Iron/sulfur cluster (351 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Beta,beta-carotene-4,4'-dione (564 Da, Ligand)
- Photosystem i reaction center subunit iii (18 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem ii reaction center protein i (4 kDa, Protein from Synechocystis sp. PCC 6803)
Resolution: 3.15 Å
EM Method: Single-particle
Fitted PDBs: 8asl
Q-score: 0.518
Zhao Z, Vercellino I, Knoppova J, Sobotka R, Murray JW, Nixon PJ, Sazanov LA, Komenda J
Nat Commun (2023) 14 pp. 4681-4681 [ DOI: doi:10.1038/s41467-023-40388-6 Pubmed: 37542031 ]
- Photosystem ii protein d1 2 (38 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit psak 1 (8 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit xii (3 kDa, Protein from Synechocystis sp. PCC 6803)
- Phylloquinone (450 Da, Ligand)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- Beta,beta-caroten-4-one (550 Da, Ligand)
- Photosystem i reaction center subunit iv (8 kDa, Protein from Synechocystis sp. PCC 6803)
- Beta-carotene (536 Da, Ligand)
- Photosystem i reaction center subunit xi (16 kDa, Protein from Synechocystis sp. PCC 6803)
- Fe (ii) ion (55 Da, Ligand)
- (1r,2s)-4-{(1e,3e,5e,7e,9e,11e,13e,15e,17e)-18-[(4s)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol (568 Da, Ligand)
- Photosystem i reaction center subunit ix (4 kDa, Protein from Synechocystis sp. PCC 6803)
- Cytochrome b559 subunit alpha (9 kDa, Protein from Synechocystis sp. PCC 6803)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Photosystem ii d2 protein (37 kDa, Protein from Synechocystis sp. PCC 6803)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Photosystem i reaction center subunit viii (4 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit ii (15 kDa, Protein from Synechocystis sp. PCC 6803)
- Pheophytin a (871 Da, Ligand)
- Chlorophyll a isomer (893 Da, Ligand)
- Cytochrome b559 subunit beta (4 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i p700 chlorophyll a apoprotein a2 (81 kDa, Protein from Synechocystis sp. PCC 6803)
- Rcii/psi complex class 2 (Complex from Synechocystis sp. PCC 6803)
- Photosystem i iron-sulfur center (8 kDa, Protein from Synechocystis sp. PCC 6803)
- Protoporphyrin ix containing fe (616 Da, Ligand)
- (3'r)-3'-hydroxy-beta,beta-caroten-4-one (566 Da, Ligand)
- Photosystem ii assembly lipoprotein ycf48 (37 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i p700 chlorophyll a apoprotein a1 (83 kDa, Protein from Synechocystis sp. PCC 6803)
- Beta,beta-carotene-4,4'-dione (564 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Photosystem i reaction center subunit iii (18 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem ii reaction center protein i (4 kDa, Protein from Synechocystis sp. PCC 6803)
RCII/PSI complex, focused refinement of PSI
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8asp
Q-score: 0.571
Zhao Z, Vercellino I, Knoppova J, Sobotka R, Murray JW, Nixon PJ, Sazanov LA, Komenda J
Nat Commun (2023) 14 pp. 4681-4681 [ DOI: doi:10.1038/s41467-023-40388-6 Pubmed: 37542031 ]
- Photosystem i reaction center subunit xii (3 kDa, Protein from Synechocystis sp. PCC 6803)
- Phylloquinone (450 Da, Ligand)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- Beta,beta-caroten-4-one (550 Da, Ligand)
- Photosystem i reaction center subunit iv (8 kDa, Protein from Synechocystis sp. PCC 6803)
- Beta-carotene (536 Da, Ligand)
- Photosystem i reaction center subunit xi (16 kDa, Protein from Synechocystis sp. PCC 6803)
- Rcii/psi complex (Complex from Synechocystis sp. PCC 6803)
- (1r,2s)-4-{(1e,3e,5e,7e,9e,11e,13e,15e,17e)-18-[(4s)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol (568 Da, Ligand)
- Photosystem i reaction center subunit ix (4 kDa, Protein from Synechocystis sp. PCC 6803)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Dodecyl-beta-d-maltoside (510 Da, Ligand)
- Photosystem i reaction center subunit viii (4 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit ii (15 kDa, Protein from Synechocystis sp. PCC 6803)
- Chlorophyll a isomer (893 Da, Ligand)
- Photosystem i p700 chlorophyll a apoprotein a2 (81 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i iron-sulfur center (8 kDa, Protein from Synechocystis sp. PCC 6803)
- (3'r)-3'-hydroxy-beta,beta-caroten-4-one (566 Da, Ligand)
- Beta,beta-carotene-4,4'-dione (564 Da, Ligand)
- Photosystem i p700 chlorophyll a apoprotein a1 (83 kDa, Protein from Synechocystis sp. PCC 6803)
- Iron/sulfur cluster (351 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Photosystem i reaction center subunit iii (18 kDa, Protein from Synechocystis sp. PCC 6803)
- Photosystem i reaction center subunit psak 1 (8 kDa, Protein from Synechocystis sp. PCC 6803)
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 6gve
Q-score: 0.387
McFarlane CR, Shah NR, Kabasakal BV, Echeverria B, Cotton CAR, Bubeck D, Murray JW
PNAS (2019) 116 pp. 20984-20990 [ DOI: doi:10.1073/pnas.1906722116 Pubmed: 31570616 ]
- Cp12 polypeptide (8 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Gapdh-cp12-prk complex (480 kDa, Complex)
- Glyceraldehyde-3-phosphate dehydrogenase (36 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Phosphoribulokinase (38 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Gapdh-cp12-prk complex (Complex from Thermosynechococcus elongatus BP-1)
Molecular Architecture of the 'stressosome', a signal transduction hub
Marles-Wright J, Grant T, Delumeau O, van Duinen G, Firbank SJ, Lewis PJ, Murray JW, Newman JA, Quin MB, Race PR, Rohou A, Tichelaar W, van Heel M, Lewis RJ
Science (2008) 322 pp. 92-96 [ Pubmed: 18832644 DOI: doi:10.1126/science.1159572 ]
Molecular Architecture of the 'stressosome', a signal transduction hub
Marles-Wright J, Grant T, Delumeau O, van Duinen G, Firbank SJ, Lewis PJ, Murray JW, Newman JA, Quin MB, Race PR, Rohou A, Tichelaar W, van Heel M, Lewis RJ
Science (2008) 322 pp. 92-96 [ Pubmed: 18832644 DOI: doi:10.1126/science.1159572 ]
Molecular Architecture of the 'stressosome', a signal transduction hub
Marles-Wright J, Grant T, Delumeau O, van Duinen G, Firbank SJ, Lewis PJ, Murray JW, Newman JA, Quin MB, Race PR, Rohou A, Tichelaar W, van Heel M, Lewis RJ
Science (2008) 322 pp. 92-96 [ Pubmed: 18832644 DOI: doi:10.1126/science.1159572 ]
Molecular Architecture of the 'stressosome', a signal transduction hub
Marles-Wright J, Grant T, Delumeau O, van Duinen G, Firbank SJ, Lewis PJ, Murray JW, Newman JA, Quin MB, Race PR, Rohou A, Tichelaar W, van Heel M, Lewis RJ
Science (2008) 322 pp. 92-96 [ Pubmed: 18832644 DOI: doi:10.1126/science.1159572 ]