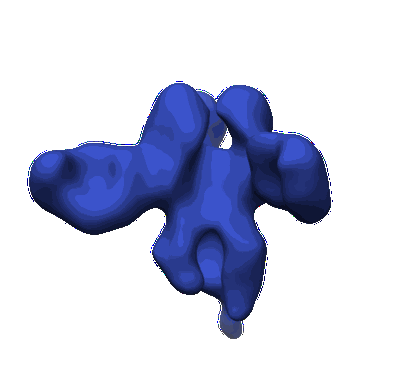
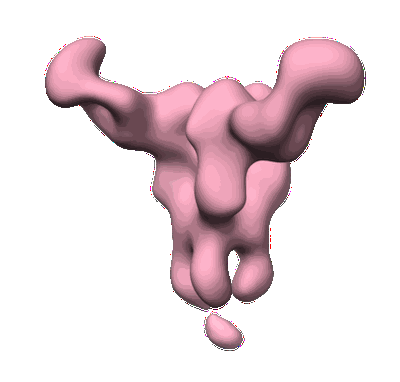CryoEM structure of monoclonal Fab 045-09 2B05 binding the lateral patch of influenza virus H1 HA
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7mem
Q-score: 0.425
Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC
Sci Transl Med (2021) 13 [ DOI: doi:10.1126/scitranslmed.abg4535 Pubmed: 34078743 ]
- Hemagglutinin ha2 chain (19 kDa, Protein from Influenza A virus (strain swl A/California/04/2009 H1N1))
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Monoclonal fab 045-09 2b05 (Complex from Homo sapiens)
- Hemagglutinin ha1 chain (36 kDa, Protein from Influenza A virus (strain swl A/California/04/2009 H1N1))
- Cryoem structure of monoclonal fab 045-09 2b05 binding the lateral patch of h1 ha (Complex)
- Heavy chain of monoclonal antibody 045-09 2b05 (12 kDa, Protein from Homo sapiens)
- Light chain of monoclonal antibody 045-09 2b05 (11 kDa, Protein from Homo sapiens)
- Hemagglutinin (Complex from Influenza A virus (strain swl A/California/04/2009 H1N1))
Negative stain map of monoclonal Fab SFV009 2G01 binding the RBS of H1 HA
Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC
Sci Transl Med (2021) 13 [ DOI: doi:10.1126/scitranslmed.abg4535 Pubmed: 34078743 ]
Negative stain map of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA
Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC
Sci Transl Med (2021) 13 [ DOI: doi:10.1126/scitranslmed.abg4535 Pubmed: 34078743 ]
Negative stain map of monoclonal Fab SFV019 2A06 binding the lateral patch of H1 HA
Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC
Sci Transl Med (2021) 13 [ DOI: doi:10.1126/scitranslmed.abg4535 Pubmed: 34078743 ]
Negative stain map of monoclonal Fab SFV015 2F02 binding the lateral patch of H1 HA
Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC
Sci Transl Med (2021) 13 [ DOI: doi:10.1126/scitranslmed.abg4535 Pubmed: 34078743 ]
Negative stain map of monoclonal Fab 047-09 4G02 binding the lateral patch of H1 HA
Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC
Sci Transl Med (2021) 13 [ DOI: doi:10.1126/scitranslmed.abg4535 Pubmed: 34078743 ]
Negative stain map of monoclonal Fab 047-09 4B06 binding the lateral patch of H1 HA
Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC
Sci Transl Med (2021) 13 [ DOI: doi:10.1126/scitranslmed.abg4535 Pubmed: 34078743 ]
Negative stain map of monoclonal Fab 23 binding the lateral patch of H1 HA
Freyn AW, Han J, Guthmiller JJ, Bailey MJ, Neu K, Turner HL, Rosado VC, Chromikova V, Huang M, Strohmeier S, Liu STH, Simon V, Krammer F, Ward AB, Palese P, Wilson PC, Nachbagauer R
PNAS (2021) 118 [ DOI: doi:10.1073/pnas.2018102118 Pubmed: 33593910 ]
Negative stain map of monoclonal Fab 45 binding the esterase domain of H1 HA
Freyn AW, Han J, Guthmiller JJ, Bailey MJ, Neu K, Turner HL, Rosado VC, Chromikova V, Huang M, Strohmeier S, Liu STH, Simon V, Krammer F, Ward AB, Palese P, Wilson PC, Nachbagauer R
PNAS (2021) 118 [ DOI: doi:10.1073/pnas.2018102118 Pubmed: 33593910 ]
Negative stain map of monoclonal Fab 50 binding the lateral patch of H1 HA
Freyn AW, Han J, Guthmiller JJ, Bailey MJ, Neu K, Turner HL, Rosado VC, Chromikova V, Huang M, Strohmeier S, Liu STH, Simon V, Krammer F, Ward AB, Palese P, Wilson PC, Nachbagauer R
PNAS (2021) 118 [ DOI: doi:10.1073/pnas.2018102118 Pubmed: 33593910 ]