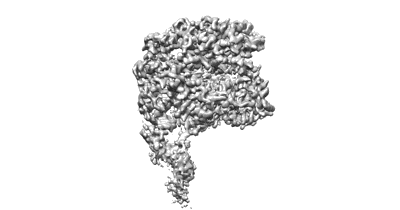
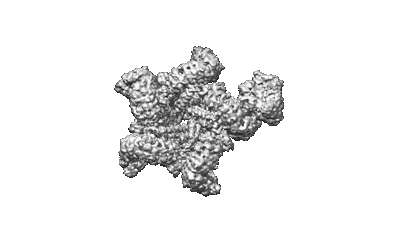
Cryo-EM structure of the prokaryotic SPARSA system complex
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8jkz
Q-score: 0.475
Zhen X, Xu X, Ye L, Xie S, Huang Z, Yang S, Wang Y, Li J, Long F, Ouyang S
Nat Commun (2024) 15 pp. 450-450 [ Pubmed: 38200015 DOI: doi:10.1038/s41467-023-44660-7 ]
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Sir2 superfamily protein (66 kDa, Protein from Geobacter sulfurreducens)
- Piwi domain protein (53 kDa, Protein from Geobacter sulfurreducens)
- Sir2/ago (Complex from Geobacter sulfurreducens PCA)
- Ago (Complex)
- Sir2 (Complex from Geobacter sulfurreducens PCA)
Cryo-EM structure of the prokaryotic SPARSA system complex
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8jl0
Q-score: 0.547
Zhen X, Xu X, Ye L, Xie S, Huang Z, Yang S, Wang Y, Li J, Long F, Ouyang S
Nat Commun (2024) 15 pp. 450-450 [ Pubmed: 38200015 DOI: doi:10.1038/s41467-023-44660-7 ]
- Nicotinamide-adenine-dinucleotide (663 Da, Ligand)
- Piwi domain protein (53 kDa, Protein from Geobacter sulfurreducens)
- The sir2/ago/guide rna/recognition dna complex (Complex from Geobacter sulfurreducens PCA)
- Dna/guide rna duplex (Complex)
- Rna (5'-r(p*ap*up*ap*ap*up*gp*gp*up*up*up*cp*up*up*ap*gp*ap*cp*gp*up*cp*gp*u)-3') (6 kDa, RNA from synthetic construct)
- Ago (Complex)
- Dna (5'-d(p*ap*cp*gp*ap*cp*gp*tp*cp*tp*ap*ap*gp*ap*ap*ap*cp*cp*ap*tp*tp*ap*t)-3') (6 kDa, DNA from synthetic construct)
- Sir2 (Complex from Geobacter sulfurreducens PCA)
- Sir2 superfamily protein (66 kDa, Protein from Geobacter sulfurreducens)
Cryo-EM structure of Cas12g-sgRNA binary complex
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8i3q
Q-score: 0.461
Liu M, Li Z, Chen J, Lin J, Lu Q, Ye Y, Zhang H, Zhang B, Ouyang S
PLoS Genet (2023) 19 pp. e1010930-e1010930 [ Pubmed: 37729124 DOI: doi:10.1371/journal.pgen.1010930 ]
- Rna (Complex)
- Zinc ion (65 Da, Ligand)
- Rna (139-mer) (44 kDa, RNA from Escherichia coli)
- Cas12g (Complex)
- Cas12g-sgrna binary complex (Complex from Escherichia coli)
- Cas12g (89 kDa, Protein from Escherichia coli)
Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 7yg7
Q-score: 0.473
Wang ZX, Liu B, Yang T, Yu D, Zhang C, Zheng L, Xie J, Liu B, Liu M, Peng H, Lai L, Ouyang Q, Ouyang S, Zhang YA
J Virol (2023) 97 pp. e0182922-e0182922 [ DOI: doi:10.1128/jvi.01829-22 Pubmed: 36943056 ]
- Rna (Complex)
- Rna (99-mer) (30 kDa, RNA from Trichoplusia ni)
- Nucleoprotein (46 kDa, Protein from Sprivivirus cyprinus)
- Ribonucleoprotein complex of the spring viraemia carp virus (Complex from Sprivirus cyprinus)
- Nucleoprotein (Complex from Trichoplusia ni)
Cryo-EM structure of Sr35 resistosome induced by AvrSr35 R381A
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7xx2
Q-score: 0.418
Zhao YB, Liu MX, Chen TT, Ma X, Li ZK, Zheng Z, Zheng SR, Chen L, Li YZ, Tang LR, Chen Q, Wang P, Ouyang S
Sci Adv (2022) 8 pp. eabq5108-eabq5108 [ Pubmed: 36083908 DOI: doi:10.1126/sciadv.abq5108 ]
- Cnl9 (105 kDa, Protein from Triticum monococcum)
- Sr35 resistosome (Complex)
- Avrsr35 (Complex from Puccinia graminis f.sp.tritici)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Sr35 (Complex from Triticum monococcum)
- Avrsr35 (66 kDa, Protein from Puccinia graminis f. sp. tritici)
Cryo-EM structure of plant NLR Sr35 resistosome
Resolution: 3.33 Å
EM Method: Single-particle
Fitted PDBs: 7xe0
Q-score: 0.351
Zhao YB, Liu MX, Chen TT, Ma X, Li ZK, Zheng Z, Zheng SR, Chen L, Li YZ, Tang LR, Chen Q, Wang P, Ouyang S
Sci Adv (2022) 8 pp. eabq5108-eabq5108 [ Pubmed: 36083908 DOI: doi:10.1126/sciadv.abq5108 ]
- Avrsr35 (Complex from Puccinia graminis f. sp. tritici)
- Sr35 resistosome (Complex)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Sr35 (105 kDa, Protein from Triticum monococcum)
- Sr35 (Complex from Triticum monococcum)
- Avrsr35 (66 kDa, Protein from Puccinia graminis f. sp. tritici)
Cryo-EM structure of binary complex of plant NLR Sr35 and effector AvrSr35
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7xvg
Q-score: 0.403
Zhao YB, Liu MX, Chen TT, Ma X, Li ZK, Zheng Z, Zheng SR, Chen L, Li YZ, Tang LR, Chen Q, Wang P, Ouyang S
Sci Adv (2022) 8 pp. eabq5108-eabq5108 [ Pubmed: 36083908 DOI: doi:10.1126/sciadv.abq5108 ]
- Avrsr35 (Complex from Puccinia graminis f. sp. tritici)
- Avrsr35 (51 kDa, Protein from Puccinia graminis f. sp. tritici)
- Sr35 (105 kDa, Protein from Triticum monococcum)
- Sr35 (Complex from Triticum monococcum)
- Binary complex of sr35 and avrsr35 (Complex)
Cryo-EM structure of SADS-CoV spike
Resolution: 3.55 Å
EM Method: Single-particle
Fitted PDBs: 6m39
Q-score: 0.44
Guan H, Wang Y, Perculija V, Saeed AFUH, Liu Y, Li J, Jan SS, Li Y, Zhu P, Ouyang S
J Virol (2020) 94 [ Pubmed: 32817223 DOI: doi:10.1128/JVI.01301-20 ]
cryo-EM structure of human PA200
Resolution: 3.75 Å
EM Method: Single-particle
Fitted PDBs: 6kwx
Q-score: 0.357
Guan H, Wang Y, Yu T, Huang Y, Li M, Saeed AFUH, Perculija V, Li D, Xiao J, Wang D, Zhu P, Ouyang S
PLoS Biol (2020) 18 pp. e3000654-e3000654 [ DOI: doi:10.1371/journal.pbio.3000654 Pubmed: 32134919 ]
- [(1~{s},2~{r},3~{r},4~{s},5~{s},6~{r})-2-[oxidanyl(phosphonooxy)phosphoryl]oxy-3,4,5,6-tetraphosphonooxy-cyclohexyl] phosphono hydrogen phosphate (819 Da, Ligand)
- Proteasome activator complex subunit 4 (215 kDa, Protein from Homo sapiens)
- Human pa200 (Cell from Homo sapiens)
- Inositol hexakisphosphate (660 Da, Ligand)