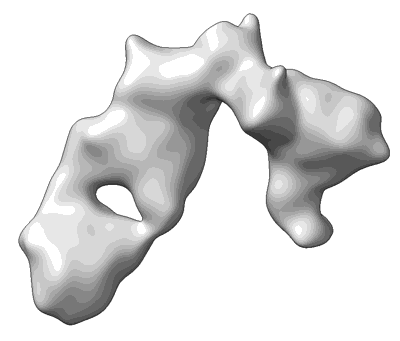
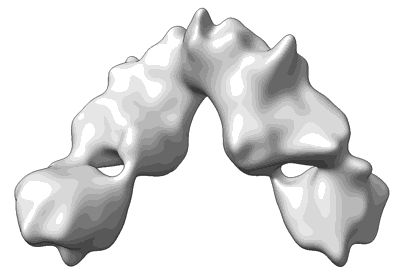
SFTSV Gn in complex with JK-8/12 Fab
Resolution: 2.37 Å
EM Method: Single-particle
Fitted PDBs: 8zhq
Q-score: 0.505
Zhang S, Shang H, Han S, Li J, Peng X, Wu Y, Yang X, Leng Y, Wang F, Cui N, Xu L, Zhang H, Guo Y, Xu X, Zhang N, Liu W, Li H
Ebiomedicine (2024) 111 pp. 105481-105481 [ DOI: doi:10.1016/j.ebiom.2024.105481 Pubmed: 39644769 ]
- Ternary complex of sftsv gn with neutralizing antibodies fab jk-8 and jk-12 (150 kDa, Complex from Homo sapiens)
- Jk-12 fab heavy chain (22 kDa, Protein from Homo sapiens)
- Jk-8 fab light chain (23 kDa, Protein from Homo sapiens)
- Envelopment polyprotein (38 kDa, Protein from Severe fever with thrombocytopenia syndrome virus)
- Jk-8 fab heavy chain (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Jk-12 fab light chain (22 kDa, Protein from Homo sapiens)
Structural basis for the nucleosome binding and chromatin compaction by the linker histone H5
Structural basis for the nucleosome binding and chromatin compaction by the linker histone H5
Structural basis for the linker histone H5-nucleosome binding and chromatin compaction
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8xjv
Q-score: 0.424
Li W, Hu J, Song F, Yu J, Peng X, Zhang S, Wang L, Hu M, Liu JC, Wei Y, Xiao X, Li Y, Li D, Wang H, Zhou BR, Dai L, Mou Z, Zhou M, Zhang H, Zhou Z, Zhang H, Bai Y, Zhou JQ, Li W, Li G, Zhu P
Cell Res (2024) 34 pp. 707-724 [ DOI: doi:10.1038/s41422-024-01009-z Pubmed: 39103524 ]
- Dna (651 kDa, DNA from synthetic construct)
- Histone h2a (14 kDa, Protein from Xenopus laevis)
- Histone h3.1 (Complex from Xenopus laevis)
- Histone h4 (Complex from Xenopus laevis)
- Histone h2a (Complex from Xenopus laevis)
- Histone h2b (Complex from Xenopus laevis)
- 12x177bp-h5 chromatin (Complex)
- Histone h5 (21 kDa, Protein from Gallus gallus)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Linker histone h5 (Complex from Gallus gallus)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Dna (660 kDa, DNA from synthetic construct)
- Widom 601 dna (Complex from synthetic construct)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
Cryo-EM structure of SARS-CoV-2 prototype-Beta chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 Delta-Omicron chimeric RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265- [ DOI: doi:10.1016/j.cell.2022.04.029 ]
Cryo-EM structure of SARS-CoV-2 prototype RBD-dimer bound to CB6 Fab
Xu K, Gao P, Liu S, Lu S, Lei W, Zheng T, Liu X, Xie Y, Zhao Z, Guo S, Tang C, Yang Y, Yu W, Wang J, Zhou Y, Huang Q, Liu C, An Y, Zhang R, Han Y, Duan M, Wang S, Yang C, Wu C, Liu X, She G, Liu Y, Zhao X, Xu K, Qi J, Wu G, Peng X, Dai L, Wang P, Gao GF
Cell (2022) 185 pp. 2265-2278.e14 [ Pubmed: 35568034 DOI: doi:10.1016/j.cell.2022.04.029 ]
SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up)
Resolution: 6.21 Å
EM Method: Single-particle
Fitted PDBs: 8hc2
Q-score: 0.154
Yu H, Liu B, Zhang Y, Gao X, Wang Q, Xiang H, Peng X, Xie C, Wang Y, Hu P, Shi J, Shi Q, Zheng P, Feng C, Tang G, Liu X, Guo L, Lin X, Li J, Liu C, Huang Y, Yang N, Chen Q, Li Z, Su M, Yan Q, Pei R, Chen X, Liu L, Hu F, Liang D, Ke B, Ke C, Li F, He J, Wang M, Chen L, Xiong X, Tang X
Nat Commun (2023) 14 pp. 1058-1058 [ Pubmed: 36828833 DOI: doi:10.1038/s41467-023-36761-0 ]
- Sars-cov-2 omicron ba.1 spike trimer (Complex from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron ba.1 spike trimer with one rbd up in complex with one yb9-258 fab (Complex)
- Yb9-258 fab (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (138 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Light chain of yb9-258 fab (23 kDa, Protein from Homo sapiens)
- Heavy chain of yb9-258 fab (23 kDa, Protein from Homo sapiens)
SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 2 YB9-258 Fabs (2 RBD up)
Resolution: 4.35 Å
EM Method: Single-particle
Fitted PDBs: 8hc3
Q-score: 0.224
Yu H, Liu B, Zhang Y, Gao X, Wang Q, Xiang H, Peng X, Xie C, Wang Y, Hu P, Shi J, Shi Q, Zheng P, Feng C, Tang G, Liu X, Guo L, Lin X, Li J, Liu C, Huang Y, Yang N, Chen Q, Li Z, Su M, Yan Q, Pei R, Chen X, Liu L, Hu F, Liang D, Ke B, Ke C, Li F, He J, Wang M, Chen L, Xiong X, Tang X
Nat Commun (2023) 14 pp. 1058-1058 [ Pubmed: 36828833 DOI: doi:10.1038/s41467-023-36761-0 ]
- Heavy chain of yb9-258 (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 omicron ba.1 spike trimer (Complex from Severe acute respiratory syndrome coronavirus 2)
- Light chain of yb9-258 (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 omicron ba.1 spike trimer with two rbd up in complex with two yb9-258 fab (Complex)
- Yb9-258 fab (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (138 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)