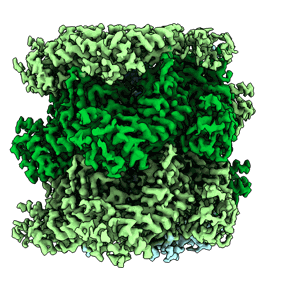
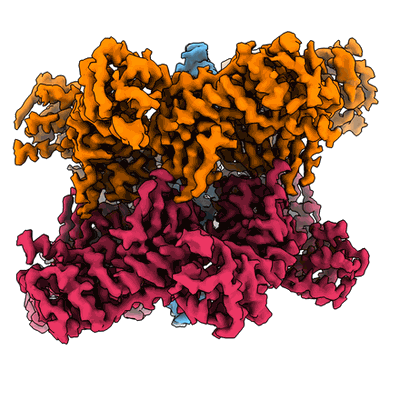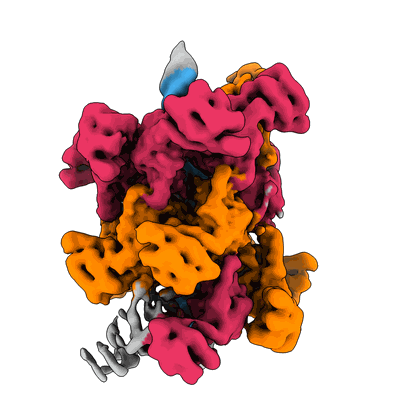CryoEM structure of TnsC(1-503) bound to TnsD(1-318) from E.coli Tn7
Resolution: 3.57 Å
EM Method: Single-particle
Fitted PDBs: 8glu
Q-score: 0.404
Shen Y, Krishnan SS, Petassi MT, Hancock MA, Peters JE, Guarne A
Mol Cell (2024) 84 pp. 2368-2381.e6 [ Pubmed: 38834067 DOI: doi:10.1016/j.molcel.2024.05.012 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Transposon tn7 transposition protein tnsc (59 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Transposon tn7 transposition protein tnsd (36 kDa, Protein from Escherichia coli)
- Complex of tnsc(1-503) bound to tnsd(1-318) with cofactors atp and adp (220 kDa, Complex from Escherichia coli)
Resolution: 3.51 Å
EM Method: Single-particle
Fitted PDBs: 8glw
Q-score: 0.399
Shen Y, Krishnan SS, Petassi MT, Hancock MA, Peters JE, Guarne A
Mol Cell (2024) 84 pp. 2368-2381.e6 [ Pubmed: 38834067 DOI: doi:10.1016/j.molcel.2024.05.012 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (50-mer) (15 kDa, DNA from Escherichia coli)
- Complex of tnsc(1-503) bound to tnsd(1-318) and dna with cofactors atp and adp (550 kDa, Complex from Escherichia coli)
- Transposon tn7 transposition protein tnsc (59 kDa, Protein from Escherichia coli)
- Transposon tn7 transposition protein tnsd (36 kDa, Protein from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Dna (50-mer) (15 kDa, DNA from Escherichia coli)
Resolution: 3.88 Å
EM Method: Single-particle
Fitted PDBs: 8glx
Q-score: 0.327
Shen Y, Krishnan SS, Petassi MT, Hancock MA, Peters JE, Guarne A
Mol Cell (2024) 84 pp. 2368-2381.e6 [ Pubmed: 38834067 DOI: doi:10.1016/j.molcel.2024.05.012 ]
- Complex of tnsc(1-503) bound to tnsd(1-318) and dna with cofactors atp and adp (500 kDa, Complex)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna (50-mer) (15 kDa, DNA from Escherichia coli)
- Tnsc(1-503) (Complex from Escherichia coli)
- Transposon tn7 transposition protein tnsc (59 kDa, Protein from Escherichia coli)
- Dna (Complex from Escherichia coli)
- Tnsd(1-318) (Complex from Escherichia coli)
- Transposon tn7 transposition protein tnsd (36 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Dna (50-mer) (15 kDa, DNA from Escherichia coli)
Resolution: 3.32 Å
EM Method: Single-particle
Fitted PDBs: 8vcj
Q-score: 0.456
Shen Y, Krishnan SS, Petassi MT, Hancock MA, Peters JE, Guarne A
Mol Cell (2024) 84 pp. 2368-2381.e6 [ Pubmed: 38834067 DOI: doi:10.1016/j.molcel.2024.05.012 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
- Complex of tnsc(1-503) bound to tnsd(1-318) and dna with cofactors atpgs and adp (550 kDa, Complex)
- Dna (15 kDa, DNA from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Transposon tn7 transposition protein tnsc (59 kDa, Protein from Escherichia coli)
- Dna (Complex from Escherichia coli)
- Transposon tn7 transposition protein tnsd (36 kDa, Protein from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Tnsc(1-503) and tnsd(1-318) (Complex from Escherichia coli)
- Dna (15 kDa, DNA from Escherichia coli)
Resolution: 3.83 Å
EM Method: Single-particle
Fitted PDBs: 8vct
Q-score: 0.396
Shen Y, Krishnan SS, Petassi MT, Hancock MA, Peters JE, Guarne A
Mol Cell (2024) 84 pp. 2368-2381.e6 [ Pubmed: 38834067 DOI: doi:10.1016/j.molcel.2024.05.012 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
- Dna (50-mer) (15 kDa, DNA from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Transposon tn7 transposition protein tnsc (59 kDa, Protein from Escherichia coli)
- Cyoem structure of the tnsc(1-503)-tnsd(1-318)-dna complex in a 6:2:1 stoichiometry from e. coli tn7 bound to atpgs and adp (Complex)
- Dna (Complex from Escherichia coli)
- Transposon tn7 transposition protein tnsd (36 kDa, Protein from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Tnsc(1-503)-tnsd(1-318) (Complex from Escherichia coli)
- Dna (50-mer) (15 kDa, DNA from Escherichia coli)
Cryo-electron microscopy structure of TnsC(1-503)A225V bound to DNA
Resolution: 3.56 Å
EM Method: Single-particle
Fitted PDBs: 7mcs
Q-score: 0.439
Shen Y, Gomez-Blanco J, Petassi MT, Peters JE, Ortega J, Guarne A
Nat Struct Mol Biol (2022) 29 pp. 143-151 [ Pubmed: 35173349 DOI: doi:10.1038/s41594-022-00724-8 ]
- Dna (5'-d(p*gp*cp*gp*cp*gp*cp*gp*cp*gp*cp*gp*cp*gp*cp*g)-3') (4 kDa, DNA from synthetic construct)
- Dna (5'-d(p*cp*gp*cp*gp*cp*gp*cp*gp*cp*gp*cp*gp*cp*gp*c)-3') (4 kDa, DNA from synthetic construct)
- Complex of tnsc bound to dna in the presence of amppnp. (30 kDa, Complex from Escherichia coli)
- Transposon tn7 transposition protein tnsc (59 kDa, Protein from Escherichia coli)
- Tnsc(1-503)a225v bound to dna (Complex from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
ATPgS bound TnsC filament from shCAST system
Resolution: 3.2 Å
EM Method: Helical reconstruction
Fitted PDBs: 7m99
Q-score: 0.477
Park JU, Tsai AW, Mehrotra E, Petassi MT, Hsieh SC, Ke A, Peters JE, Kellogg EH
Science (2021) 373 pp. 768-774 [ Pubmed: 34385391 DOI: doi:10.1126/science.abi8976 ]
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
- Tnsc (31 kDa, Protein from Scytonema hofmannii)
- Dna (5'-d(p*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*ap*a)-3') (6 kDa, DNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Dna (5'-d(p*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*tp*t)-3') (6 kDa, DNA from synthetic construct)
- Helical filament of tnsc monomers bound to dna (45 MDa, Complex from Scytonema hofmannii)
ADP-AlF3 bound TnsC structure from ShCAST system
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7m9a
Q-score: 0.342
Park JU, Tsai AW, Mehrotra E, Petassi MT, Hsieh SC, Ke A, Peters JE, Kellogg EH
Science (2021) 373 pp. 768-774 [ Pubmed: 34385391 DOI: doi:10.1126/science.abi8976 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Tnsc (31 kDa, Protein from Scytonema hofmannii)
- Head-to-head dimer of tnsc hexamers bound to dna (400 kDa, Complex from Scytonema hofmannii)
- Dna (27-mer) (8 kDa, DNA from synthetic construct)
- Dna (27-mer) (8 kDa, DNA from synthetic construct)
ADP-AlF3 bound TnsC structure in closed form
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 7m9b
Q-score: 0.317
Park JU, Tsai AW, Mehrotra E, Petassi MT, Hsieh SC, Ke A, Peters JE, Kellogg EH
Science (2021) 373 pp. 768-774 [ Pubmed: 34385391 DOI: doi:10.1126/science.abi8976 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Tnsc (31 kDa, Protein from Scytonema hofmannii)
- Head to head dimer of tnsc hexamers bound to dna (400 kDa, Complex from Scytonema hofmannii)
- Dna (27-mer) (8 kDa, DNA from synthetic construct)
- Dna (27-mer) (8 kDa, DNA from synthetic construct)
ADP-AlF3 bound TnsC structure in open form
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 7m9c
Q-score: 0.286
Park JU, Tsai AW, Mehrotra E, Petassi MT, Hsieh SC, Ke A, Peters JE, Kellogg EH
Science (2021) 373 pp. 768-774 [ Pubmed: 34385391 DOI: doi:10.1126/science.abi8976 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Dna (34-mer) (10 kDa, DNA from synthetic construct)
- Tnsc (31 kDa, Protein from Scytonema hofmannii)
- Dna (34-mer) (10 kDa, DNA from synthetic construct)
- Head to head dimer of tnsc hexameters bound to dna (400 kDa, Complex from Scytonema hofmannii)