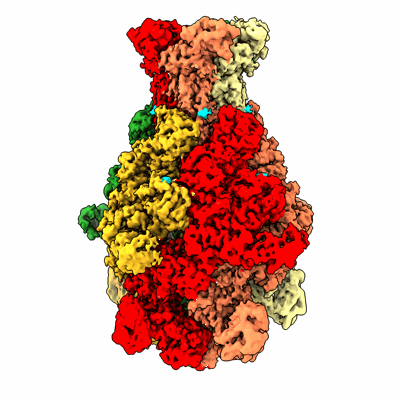
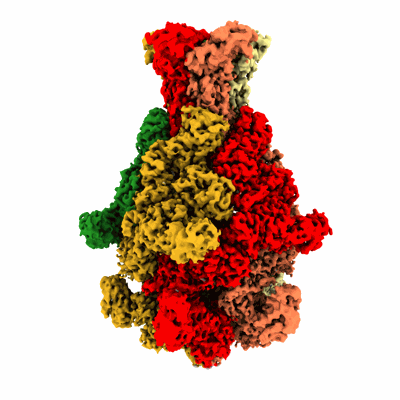
Rat alpha5beta1 integrin, headpiece
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 8oxz
Q-score: 0.293
Shafaq-Zadah M, Dransart E, Wunder C, Chambon V, Valades-Cruz CA, Leconte L, Sarangi NK, Robinson J, Bai S-K, Regmi R, Di Cicco A, Hovasse A, Bartels R, Nilsson UJ, Cianferani-Sanglier S, Leffler H, Keyes TE, Levy D, Raunser S, Roderer D, Johannes L
To Be Published
- Integrin beta-1 (88 kDa, Protein from Rattus norvegicus)
- Integrin subunit alpha 5 (119 kDa, Protein from Rattus norvegicus)
- Alpha5beta1 integrin from rattus norvegicus (Complex from Rattus norvegicus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8cq0
Q-score: 0.466
Ng'ang'a PN, Folz J, Kucher S, Roderer D, Xu Y, Sitsel O, Belyy A, Prumbaum D, Kuhnemuth R, Assafa TE, Dong M, Seidel CAM, Bordignon E, Raunser S
Sci Adv (2025) 11 pp. eadr2019-eadr2019 [ Pubmed: 39752508 DOI: doi:10.1126/sciadv.adr2019 ]
- Tcda1 (285 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (1 MDa, Complex from Photorhabdus luminescens)
Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8cq2
Q-score: 0.448
Ng'ang'a PN, Folz J, Kucher S, Roderer D, Xu Y, Sitsel O, Belyy A, Prumbaum D, Kuhnemuth R, Assafa TE, Dong M, Seidel CAM, Bordignon E, Raunser S
Sci Adv (2025) 11 pp. eadr2019-eadr2019 [ Pubmed: 39752508 DOI: doi:10.1126/sciadv.adr2019 ]
- Tcda1 (285 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (1 MDa, Complex from Photorhabdus luminescens)
Structure of P. luminescens TccC3-F-actin complex
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 7z7h
Q-score: 0.366
Belyy A, Lindemann F, Roderer D, Funk J, Bardiaux B, Protze J, Bieling P, Oschkinat H, Raunser S
Nat Commun (2022) 13 pp. 4202-4202 [ Pubmed: 35858890 DOI: doi:10.1038/s41467-022-31836-w ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Nicotinamide (122 Da, Ligand)
- Structure of p. luminescence tccc3-f-actin complex (Complex from Photorhabdus luminescens)
- Magnesium ion (24 Da, Ligand)
- Actin, alpha skeletal muscle (42 kDa, Protein from rabbit)
- Adenosine-5-diphosphoribose (559 Da, Ligand)
- Maltose/maltodextrin-binding periplasmic protein,tccc3 (63 kDa, Protein from Photorhabdus luminescens)
Structure of ADP-ribosylated F-actin
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7z7i
Q-score: 0.444
Belyy A, Lindemann F, Roderer D, Funk J, Bardiaux B, Protze J, Bieling P, Oschkinat H, Raunser S
Nat Commun (2022) 13 pp. 4202-4202 [ Pubmed: 35858890 DOI: doi:10.1038/s41467-022-31836-w ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Structure of adp-ribosylated f-actin (Complex from Oryctolagus cuniculus)
- Magnesium ion (24 Da, Ligand)
- Actin, alpha skeletal muscle (42 kDa, Protein from rabbit)
- Adenosine-5-diphosphoribose (559 Da, Ligand)
Morganella morganii TcdA4 in complex with porcine mucosa heparin
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 6yew
Q-score: 0.478
Roderer D, Brocker F, Sitsel O, Kaplonek P, Leidreiter F, Seeberger PH, Raunser S
Nat Commun (2020) 11 pp. 2694-2694 [ Pubmed: 32483155 DOI: doi:10.1038/s41467-020-16536-7 ]
Structure of Photorhabdus luminescens Tc holotoxin pore, Mutation TccC3-D651A
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6sue
Q-score: 0.385
Roderer D, Hofnagel O, Benz R, Raunser S
PNAS (2019) 116 pp. 23083-23090 [ DOI: doi:10.1073/pnas.1909821116 Pubmed: 31666324 ]
- Tcdb2,tccc3 (274 kDa, Protein from Photorhabdus luminescens)
- Tc holotoxin composed of the tcda1 pentamer and tcdb2-tccc3 (1 MDa, Complex from Photorhabdus luminescens)
- Tcda1 (283 kDa, Protein from Photorhabdus luminescens)
Structure of Photorhabdus luminescens Tc holotoxin pore
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6suf
Q-score: 0.396
Roderer D, Hofnagel O, Benz R, Raunser S
PNAS (2019) 116 pp. 23083-23090 [ DOI: doi:10.1073/pnas.1909821116 Pubmed: 31666324 ]
- Tc holotoxin complex in the pore form, formed by the tcda1 pentamer and tcdb2-tccc3 (1 MDa, Complex from Photorhabdus luminescens)
- Tcdb2,tccc3 (274 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (283 kDa, Protein from Photorhabdus luminescens)
Cryo-EM structure of Morganella morganii TcdA4
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 6rw9
Q-score: 0.423
Leidreiter F, Roderer D, Meusch D, Gatsogiannis C, Benz R, Raunser S
Sci Adv (2019) 5 pp. eaax6497-eaax6497 [ Pubmed: 31663026 DOI: doi:10.1126/sciadv.aax6497 ]
Cryo-EM structure of Yersinia pseudotuberculosis TcaA-TcaB
Resolution: 3.25 Å
EM Method: Single-particle
Fitted PDBs: 6rwb
Q-score: 0.451
Leidreiter F, Roderer D, Meusch D, Gatsogiannis C, Benz R, Raunser S
Sci Adv (2019) 5 pp. eaax6497-eaax6497 [ Pubmed: 31663026 DOI: doi:10.1126/sciadv.aax6497 ]
- Y. pseudotuberculosis tcaa-tcab pentamer (1 MDa, Complex from Yersinia pseudotuberculosis)
- Toxin,toxin complex subunit tcab,putative toxin subunit,putative toxin subunit,toxin,toxin complex subunit tcab,putative toxin subunit,putative toxin subunit,toxin,toxin complex subunit tcab,putative toxin subunit,putative toxin subunit,toxin,toxin complex subunit tcab,putative toxin subunit,putative toxin subunit (231 kDa, Protein from Yersinia pseudotuberculosis)