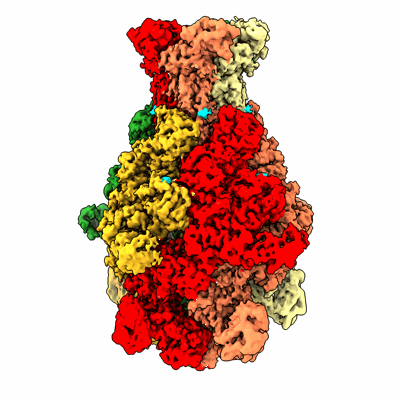
Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8cq0
Q-score: 0.466
Ng'ang'a PN, Folz J, Kucher S, Roderer D, Xu Y, Sitsel O, Belyy A, Prumbaum D, Kuhnemuth R, Assafa TE, Dong M, Seidel CAM, Bordignon E, Raunser S
Sci Adv (2025) 11 pp. eadr2019-eadr2019 [ Pubmed: 39752508 DOI: doi:10.1126/sciadv.adr2019 ]
- Tcda1 (285 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (1 MDa, Complex from Photorhabdus luminescens)
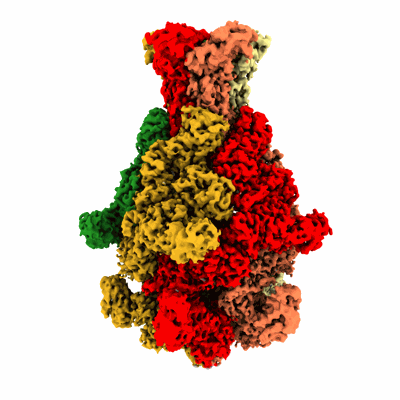
Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8cq2
Q-score: 0.448
Ng'ang'a PN, Folz J, Kucher S, Roderer D, Xu Y, Sitsel O, Belyy A, Prumbaum D, Kuhnemuth R, Assafa TE, Dong M, Seidel CAM, Bordignon E, Raunser S
Sci Adv (2025) 11 pp. eadr2019-eadr2019 [ Pubmed: 39752508 DOI: doi:10.1126/sciadv.adr2019 ]
- Tcda1 (285 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (1 MDa, Complex from Photorhabdus luminescens)
Structure of P. luminescens TccC3-F-actin complex
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 7z7h
Q-score: 0.366
Belyy A, Lindemann F, Roderer D, Funk J, Bardiaux B, Protze J, Bieling P, Oschkinat H, Raunser S
Nat Commun (2022) 13 pp. 4202-4202 [ Pubmed: 35858890 DOI: doi:10.1038/s41467-022-31836-w ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Nicotinamide (122 Da, Ligand)
- Structure of p. luminescence tccc3-f-actin complex (Complex from Photorhabdus luminescens)
- Magnesium ion (24 Da, Ligand)
- Actin, alpha skeletal muscle (42 kDa, Protein from rabbit)
- Adenosine-5-diphosphoribose (559 Da, Ligand)
- Maltose/maltodextrin-binding periplasmic protein,tccc3 (63 kDa, Protein from Photorhabdus luminescens)
Structure of ADP-ribosylated F-actin
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7z7i
Q-score: 0.444
Belyy A, Lindemann F, Roderer D, Funk J, Bardiaux B, Protze J, Bieling P, Oschkinat H, Raunser S
Nat Commun (2022) 13 pp. 4202-4202 [ Pubmed: 35858890 DOI: doi:10.1038/s41467-022-31836-w ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Structure of adp-ribosylated f-actin (Complex from Oryctolagus cuniculus)
- Magnesium ion (24 Da, Ligand)
- Actin, alpha skeletal muscle (42 kDa, Protein from rabbit)
- Adenosine-5-diphosphoribose (559 Da, Ligand)
Structure of Photorhabdus luminescens Tc holotoxin pore, Mutation TccC3-D651A
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6sue
Q-score: 0.385
Roderer D, Hofnagel O, Benz R, Raunser S
PNAS (2019) 116 pp. 23083-23090 [ DOI: doi:10.1073/pnas.1909821116 Pubmed: 31666324 ]
- Tcdb2,tccc3 (274 kDa, Protein from Photorhabdus luminescens)
- Tc holotoxin composed of the tcda1 pentamer and tcdb2-tccc3 (1 MDa, Complex from Photorhabdus luminescens)
- Tcda1 (283 kDa, Protein from Photorhabdus luminescens)
Structure of Photorhabdus luminescens Tc holotoxin pore
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6suf
Q-score: 0.396
Roderer D, Hofnagel O, Benz R, Raunser S
PNAS (2019) 116 pp. 23083-23090 [ DOI: doi:10.1073/pnas.1909821116 Pubmed: 31666324 ]
- Tc holotoxin complex in the pore form, formed by the tcda1 pentamer and tcdb2-tccc3 (1 MDa, Complex from Photorhabdus luminescens)
- Tcdb2,tccc3 (274 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (283 kDa, Protein from Photorhabdus luminescens)
PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3)
Resolution: 3.95 Å
EM Method: Single-particle
Fitted PDBs: 6h6e
Q-score: 0.371
Gatsogiannis C, Merino F, Roderer D, Balchin D, Schubert E, Kuhlee A, Hayer-Hartl M, Raunser S
Nature (2018) 563 pp. 209-213 [ DOI: doi:10.1038/s41586-018-0556-6 Pubmed: 30232455 ]
- Tcdb2,tccc3 (273 kDa, Protein from Photorhabdus luminescens)
- Ptc3 holotoxin complex in prepore state (5 x tcda1, 1 x tcdb2, 1 x tccc3) (Complex from Photorhabdus luminescens)
- Tcda1 (283 kDa, Protein from Photorhabdus luminescens)
- Tcdb2/tccc3 (Complex from Photorhabdus luminescens)
- Tcda1 (Complex from Photorhabdus luminescens)
PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A
Resolution: 3.72 Å
EM Method: Single-particle
Fitted PDBs: 6h6f
Q-score: 0.375
Gatsogiannis C, Merino F, Roderer D, Balchin D, Schubert E, Kuhlee A, Hayer-Hartl M, Raunser S
Nature (2018) 563 pp. 209-213 [ DOI: doi:10.1038/s41586-018-0556-6 Pubmed: 30232455 ]
- Ptc3 holotoxin complex in prepore state - mutant tcc-d651a (1 MDa, Complex)
- Tcdb2,tccc3,tccc3 (273 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (283 kDa, Protein from Photorhabdus luminescens)
- Tcda1 (Complex from Photorhabdus luminescens)
- Tcdb2/tccc3-d651a (Complex from Photorhabdus luminescens)
Cryo-EM structure of the Tc toxin TcdA1 in its pore state
Resolution: 3.46 Å
EM Method: Single-particle
Fitted PDBs: 5lki, 5lkh
Q-score: 0.2485
Gatsogiannis C, Merino F, Prumbaum D, Roderer D, Leidreiter F, Meusch D, Raunser S
Nat Struct Mol Biol (2016) 23 pp. 884-890 [ Pubmed: 27571177 DOI: doi:10.1038/nsmb.3281 ]
- Tcda1 (283 kDa, Protein from Photorhabdus luminescens)
- Structure of tcda1 in pore state, determined in lipid nanodisc (1 MDa, Complex from Photorhabdus luminescens)