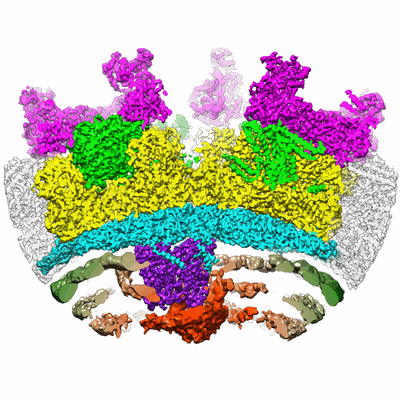
Cryo-EM structure of bluetongue virus capsid protein VP5 at low endosomal pH
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7rtn
Q-score: 0.46
Cryo-EM structure of bluetongue virus capsid protein VP5 at low endosomal pH intermediate state 2
Resolution: 3.8 Å
EM Method: Single-particle
Fitted PDBs: 7rto
Q-score: 0.402
Cryo-EM structure of bluetongue virus capsid protein VP5 at low endosomal pH intermediate state 1
Cryo-EM structure of bluetongue virus capsid protein at low endosomal pH intermediate state 3
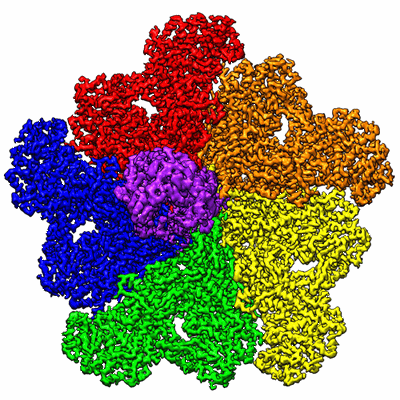
In situ structure of BTV RNA-dependent RNA polymerase in BTV virion
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6pns
Q-score: 0.484
He Y, Shivakoti S, Ding K, Cui Y, Roy P, Zhou ZH
PNAS (2019) 116 pp. 16535-16540 [ DOI: doi:10.1073/pnas.1905849116 Pubmed: 31350350 ]
- Inner core structural protein vp3 (103 kDa, Protein from Bluetongue virus 1)
- Bluetongue virus 1 (Virus from Bluetongue virus 1)
- Rna-directed rna polymerase (149 kDa, Protein from Bluetongue virus 1)
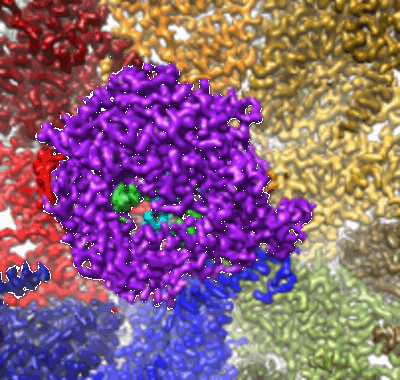
In situ structure of BTV RNA-dependent RNA polymerase in BTV core
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 6po2
Q-score: 0.497
He Y, Shivakoti S, Ding K, Cui Y, Roy P, Zhou ZH
PNAS (2019) 116 pp. 16535-16540 [ DOI: doi:10.1073/pnas.1905849116 Pubmed: 31350350 ]
- Inner core structural protein vp3 (103 kDa, Protein from Bluetongue virus 1)
- Bluetongue virus 1 (Virus from Bluetongue virus 1)
- Rna-directed rna polymerase (149 kDa, Protein from Bluetongue virus 1)
In situ structure of Rotavirus RNA-dependent RNA polymerase at duplex-open state
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6ogy
Q-score: 0.537
Ding K, Celma CC, Zhang X, Chang T, Shen W, Atanasov I, Roy P, Zhou ZH
Nat Commun (2019) 10 pp. 2216-2216 [ Pubmed: 31101900 DOI: doi:10.1038/s41467-019-10236-7 ]
- Rna-dependent rna polymerase of rotavirus a (125 kDa, Protein from Rotavirus A)
- Rotavirus a (Virus from Rotavirus A)
- Dna/rna (5'-d(*(gtg))-r(p*gp*c)-3') (1 kDa, Macromolecule from Rotavirus A)
- Rna (5'-r(p*ap*gp*cp*c)-3') (1 kDa, RNA from Rotavirus A)
- Inner capsid protein vp2 (103 kDa, Protein from Rotavirus A)
In situ structure of Rotavirus RNA-dependent RNA polymerase at transcript-elongated state
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 6ogz
Q-score: 0.517
Ding K, Celma CC, Zhang X, Chang T, Shen W, Atanasov I, Roy P, Zhou ZH
Nat Commun (2019) 10 pp. 2216-2216 [ Pubmed: 31101900 DOI: doi:10.1038/s41467-019-10236-7 ]
- Uridine 5'-triphosphate (484 Da, Ligand)
- Rna (5'-r(p*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*a)-3') (5 kDa, RNA from Rotavirus A)
- Rna-dependent rna polymerase of rotavirus a (125 kDa, Protein from Rotavirus A)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Rotavirus a (Virus from Rotavirus A)
- Rna (5'-r(p*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*ap*up*a)-3') (5 kDa, RNA from Rotavirus A)
- Inner capsid protein vp2 (103 kDa, Protein from Rotavirus A)